Visualized rapid nucleic acid testing method based on CRISPR-Cas12a system and application
A nucleic acid and nucleic acid probe technology, which can be used in biochemical equipment and methods, and the determination/inspection of microorganisms, and can solve problems such as limiting rapid detection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
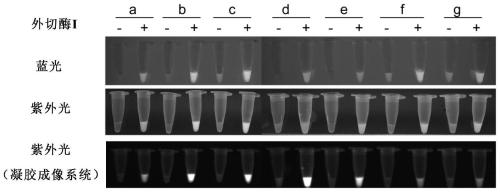
[0164] Example 1: Using exonuclease (Exonuclease I) to cut ssDNA-reporter to evaluate the influence of different base modification methods on the signal intensity of fluorescence detection
[0165] The nucleic acid detection experiment based on CRISPR-Cas12a technology is mainly divided into two steps: 1) amplification of the target nucleic acid molecule; 2) Cas12a enzyme digestion reaction: adding a non-fluorescent luminescent group at one end and a fluorescence quenching group at the other end to the system. The specific ssDNA fluorescent reporter molecule, after the Cas12 / sgRNA complex binds to the target DNA, stimulates the cleavage activity of the Cas12a protein on the ssDNA-reporter, thereby generating a free fluorescent luminescent group and emitting detectable fluorescence. In order to screen the base modification method that can enhance the signal intensity of fluorescence visualization detection, the combination method is: 5' fluorescent group-NNNNNNNNNNNNNN-3' quench...
Embodiment 2
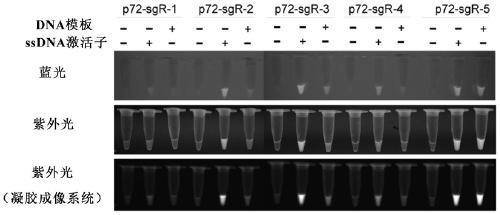
[0175] Example 2: A specific PCR primer pair and sgRNA targeting the p72 gene were designed for the nucleic acid detection of African swine fever
[0176] The p72 gene (MK333180) and genome (AY261366.1) sequences of African swine fever virus were downloaded from the NCBI database, respectively. Send the company to synthesize a partial fragment of the p72 gene and insert it into the cloning vector (pMD18T-p72). Use the primer3 software to design primer pairs for amplifying the p72 gene (Table 2); for the PCR amplification region, use the CRISPR-offinder software (https: / / sourceforge.net / projects / crispr-offinder-v1-2 / ) to design 5 primers sgRNA, PAM is TTTV, using sgRNA empty vector (pUC57-T7-sgRNA) as a template to design primer pairs for in vitro transcription (Table 2). The experimental steps for nucleic acid detection using CRISP-Cas12 are as follows:
[0177] (1) PCR amplification of target gene fragments: Prepare 50 μL of PCR reaction solution, including 25 μL of Ex taq ...
Embodiment 3
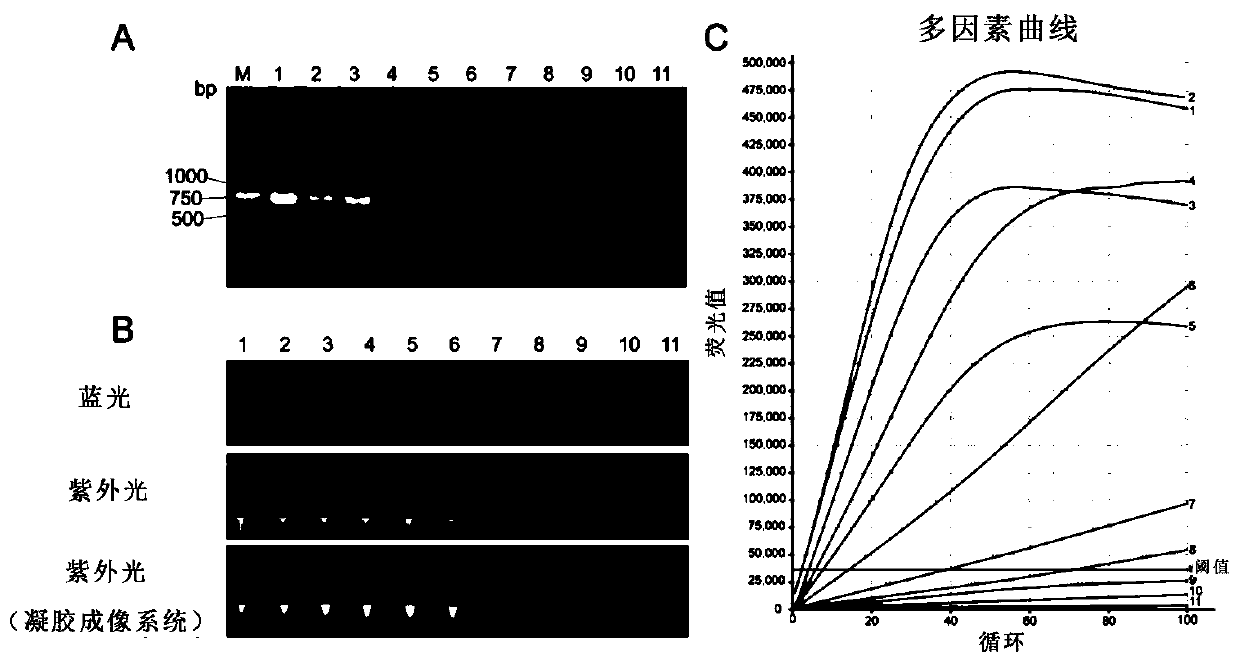
[0204] Example 3: Evaluating the sensitivity of CRISPR-Cas12a-assisted nucleic acid visualization method for detecting African swine fever virus nucleic acid
[0205]In this example, to further evaluate the sensitivity of CRISPR-Cas12a assisted nucleic acid visualization detection of African swine fever virus nucleic acid method, the pMD18T-p72 plasmid with different copy numbers was diluted as a template, and CRISPR-Cas12a nucleic acid visualization detection was performed after PCR amplification. The experimental procedure is as follows:
[0206] (1) PCR amplification: The pMD18T-p72 plasmid was diluted to 8×10 7 copy / µl, 8×10 6 copy / µl, 8×10 5 copy / µl, 8×10 4 copy / µl, 8×10 3 copy / µl, 8×10 2 copy / µl, 8×10 1 copies / µl, 4×10 1 copy / µl, 2×10 1 copy / µl, 8×10 0 Copy / microliter, etc., using plasmids of different dilutions as templates, and p72-PCR-F and p72-PCR-R as primers, PCR amplification (Table 1). The specific steps are that the reaction system is 50 μL, including ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com