Visual rapid nucleic acid detection method and application based on CRISPR-Cas12a system
A technology of target nucleic acid and nucleic acid probe, which is applied in the field of visual detection of target nucleic acid molecules and can solve problems such as limiting rapid detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
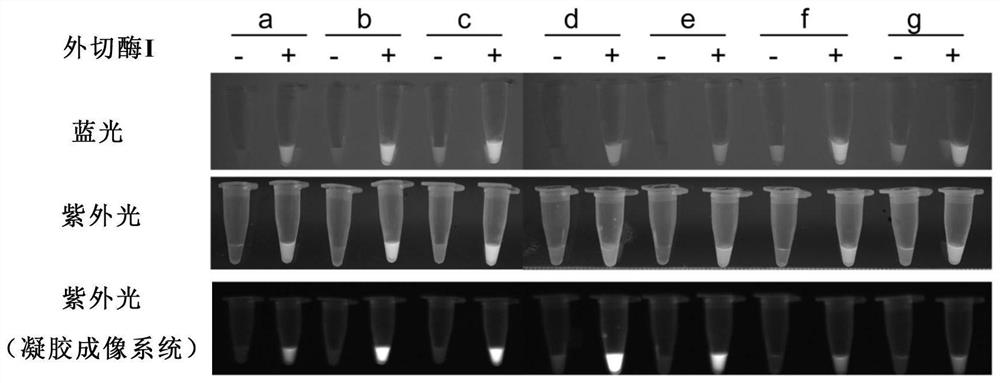
[0164] Example 1: Using exonuclease (Exonuclease I) to cut ssDNA-reporter to evaluate the influence of different base modification methods on the signal intensity of fluorescence detection
[0165] The nucleic acid detection experiment based on CRISPR-Cas12a technology is mainly divided into two steps: 1) Amplification of target nucleic acid molecules; 2) Cas12a digestion reaction: adding a non-fluorescent light-emitting group at one end and a fluorescence quenching group at the other end into the system The specific ssDNA fluorescent reporter molecule, after the Cas12 / sgRNA complex binds to the target DNA, excites the cleavage activity of Cas12a protein on ssDNA-reporter, thereby generating free fluorescent light-emitting groups and emitting detectable fluorescence. In order to screen the base modification method that can enhance the intensity of fluorescence visualization detection signal, the combination method is: 5' fluorophore-NNNNNNNNNNNN-3' quenching group, N represents...
Embodiment 2
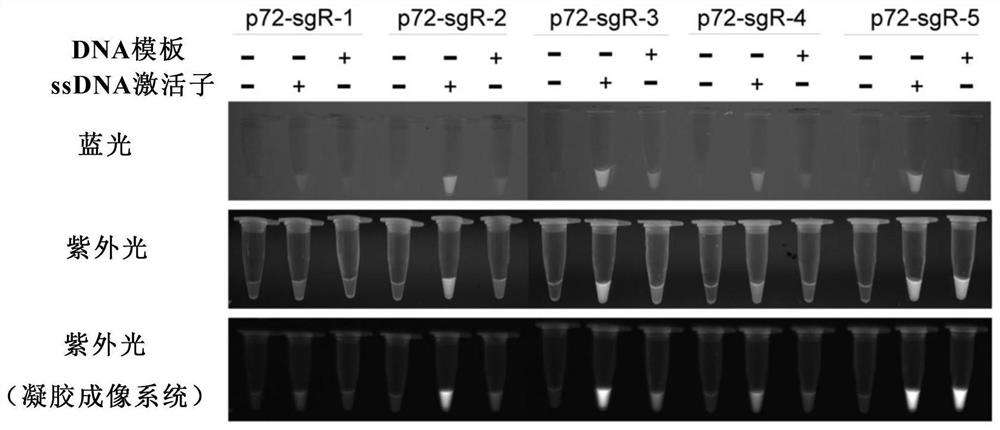
[0175] Example 2: Design of specific PCR primer pair and sgRNA targeting p72 gene for nucleic acid detection of African swine fever
[0176] The p72 gene (MK333180) and genome (AY261366.1) sequences of African swine fever virus were downloaded from the NCBI database, respectively. Send the company to synthesize a partial fragment of the p72 gene and insert it into the cloning vector (pMD18T-p72). Primer pairs for amplifying the p72 gene were designed using primer3 software (Table 2); for PCR amplification regions, 5 primers were designed using CRISPR-offinder software (https: / / sourceforge.net / projects / crispr-offinder-v1-2 / ) sgRNA, PAM is TTTV, and the primer pair for in vitro transcription was designed with the sgRNA empty vector (pUC57-T7-sgRNA) as the template (Table 2). The experimental steps for nucleic acid detection using CRISP-Cas12 are as follows:
[0177] (1) PCR amplification of target gene fragments: configure 50 μL of PCR reaction solution, including 25 μL of Ex ...
Embodiment 3
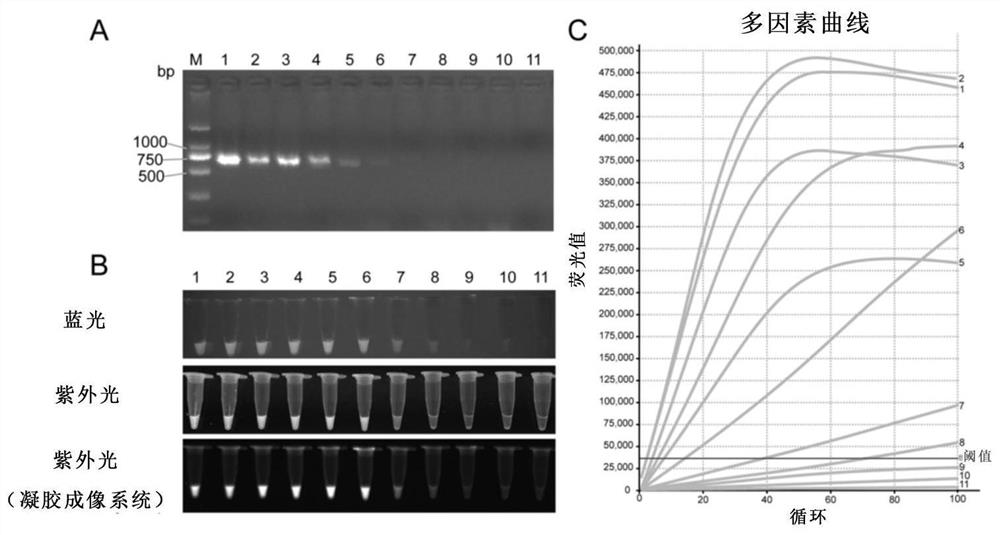
[0204] Example 3: Assessing the sensitivity of the CRISPR-Cas12a-assisted nucleic acid visualization method for detection of African swine fever virus nucleic acid
[0205]In this example, to further evaluate the sensitivity of the CRISPR-Cas12a-assisted nucleic acid visualization method for detecting African swine fever virus nucleic acid, the pMD18T-p72 plasmids with different copy numbers were diluted as templates, and CRISPR-Cas12a nucleic acid visualization was performed after PCR amplification. The experimental process is as follows:
[0206] (1) PCR amplification: fold the pMD18T-p72 plasmid to 8×10 7 copy / µl, 8×10 6 copy / µl, 8×10 5 copy / µl, 8×10 4 copy / µl, 8×10 3 copy / µl, 8×10 2 copy / µl, 8×10 1 copies / µl, 4×10 1 copy / µl, 2×10 1 copy / µl, 8×10 0 Copy / microliter, etc., using plasmids with different dilutions as templates, p72-PCR-F and p72-PCR-R as primers, PCR amplification (Table 1). The specific steps are that the reaction system is 50 μL, of which Extaq Mix ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com