InDel molecular marker closely linked with gen of rice in heading stage and application
A molecular marker and heading stage technology, applied in the field of DNA molecular marker technology and molecular biology, can solve the problem of unsaturated InDel marker, and achieve the effect of obvious band type and polymorphism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Design of InDel Molecular Marker Detection Primers Tightly Linked to Heading Date QTL in Rice
[0058] 1.1 Primer design method:
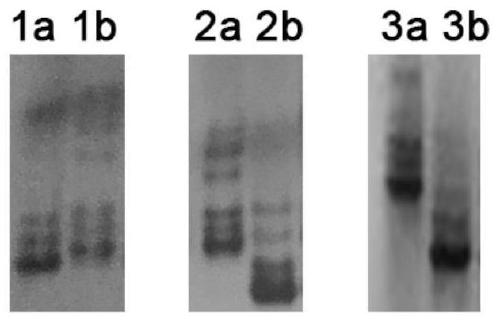
[0059] In the chromosomal segments where InDel markers need to be developed, molecular markers were designed based on the genome sequencing results of japonica rice (Nipponbare) and indica rice (9311). The sequence of the Nipponbare PAC / BAC clone in this region was downloaded from the TIGR website (The Institute of Genome Research) (http: / / www.tigr.org / tdb / rice). Select the required PAC / BAC clones, and split each clone into 2kb contiguous small fragments. Then use these 2kb sequences to compare the sequences with the indica rice variety 9311, and screen the sequences with insertion / deletion bases of 7-12 bp as candidate sequences. Based on the Nipponbare genome sequence, primers were designed within 200 bp of each flank of the screened InDel site. The primers were designed so that Primer3plus ( http: / / www.bioinformatics.nl / cgi-bin / prime...
Embodiment 2
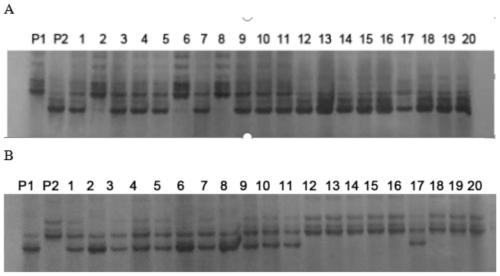
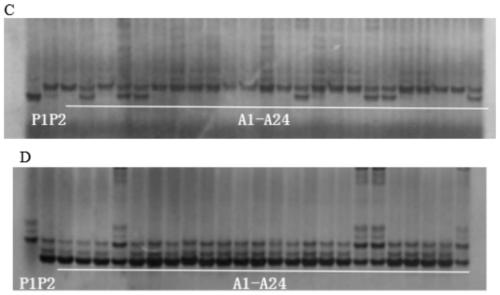
[0090] Application of Primers for InDel Molecular Markers IN2 and IN10 in Rice Genes
[0091] 2.1 Rice genomic DNA extraction
[0092] CSSL41, Shengdao 16, and 24 exchange lines (A1-A24) obtained by screening all F2 generation recessive (late heading) populations with RM527 and PSM388 as boundary markers.
[0093] The total DNA of rice genome was extracted by the improved TPS simple method, and the specific steps were as follows:
[0094] 2.1.1. At the peak tillering stage, take 1 to 2 young leaves from the upper part of each plant, and store them in a -80°C refrigerator for later use;
[0095] 2.1.2. When extracting DNA, take 2-4cm long rice leaves and put them into a 1.5ml centrifuge tube, put them in liquid nitrogen, grind them, add 900ml of TPS extract solution, and bathe in 75°C water bath for 30-60min;
[0096] 2.1.3 Centrifuge at 12000rpm for 10min, absorb about 500ml of the supernatant and transfer to a new 1.5ml centrifuge tube;
[0097] 2.1.4. Add pre-cooled isopr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com