Novel diagnostic method of acute myeloid leukemia and application thereof
A technology for acute myeloid and leukemia, which is applied in the field of tumor diagnosis and can solve problems such as difficult coverage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
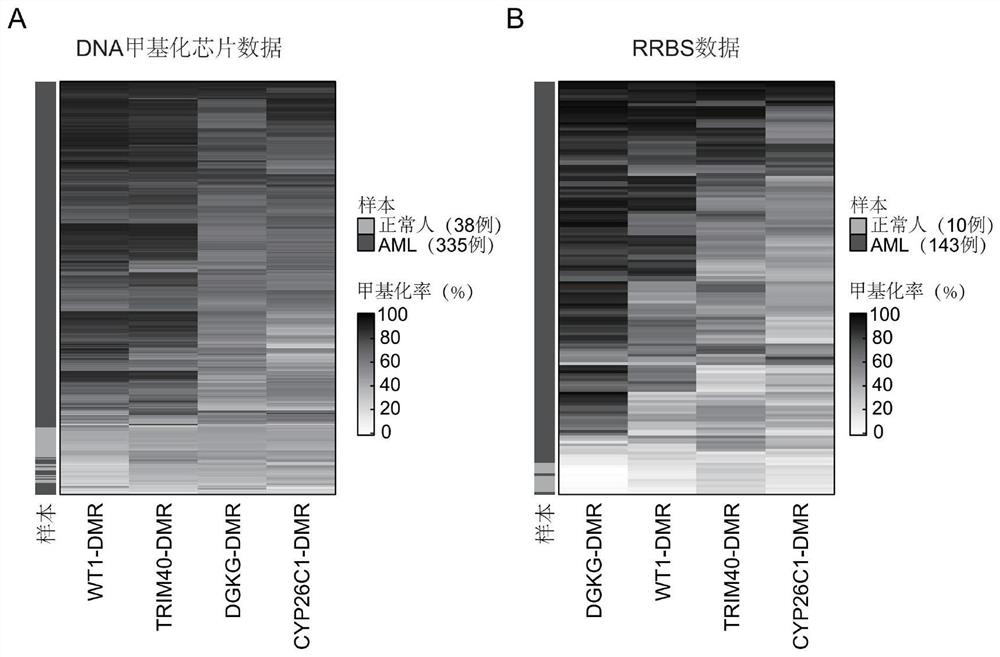
[0023] Example 1 By analyzing the DNA methylation data in TCGA and GEO databases, comparing the methylation of DNA methylation elements such as CYP26C1-DMR, DGKG-DMR, WT1-DMR, TRIM40-DMR in AML patients and normal people level difference
[0024] The present invention collects 450K DNA methylation chip data of 335 cases of AML patients and 38 cases of normal people from TCGA and GEO databases, and uses this data to analyze CYP26C1-DMR, DGKG-DMR, WT1-DMR, TRIM40-DMR, etc. 4 The methylation level of each methylation element is different in AML patients and normal people. In order to verify the correctness of this discovery, the inventors collected data from another DNA methylation detection method—genome-wide reduced methylation sequencing (RRBS), which included 143 AML patients and 10 normal individuals.
[0025] Results: Both DNA methylation microarray data and RRBS data showed that the methylation levels of four DNA methylation elements, including CYP26C1-DMR, DGKG-DMR, WT1-DM...
Embodiment 2
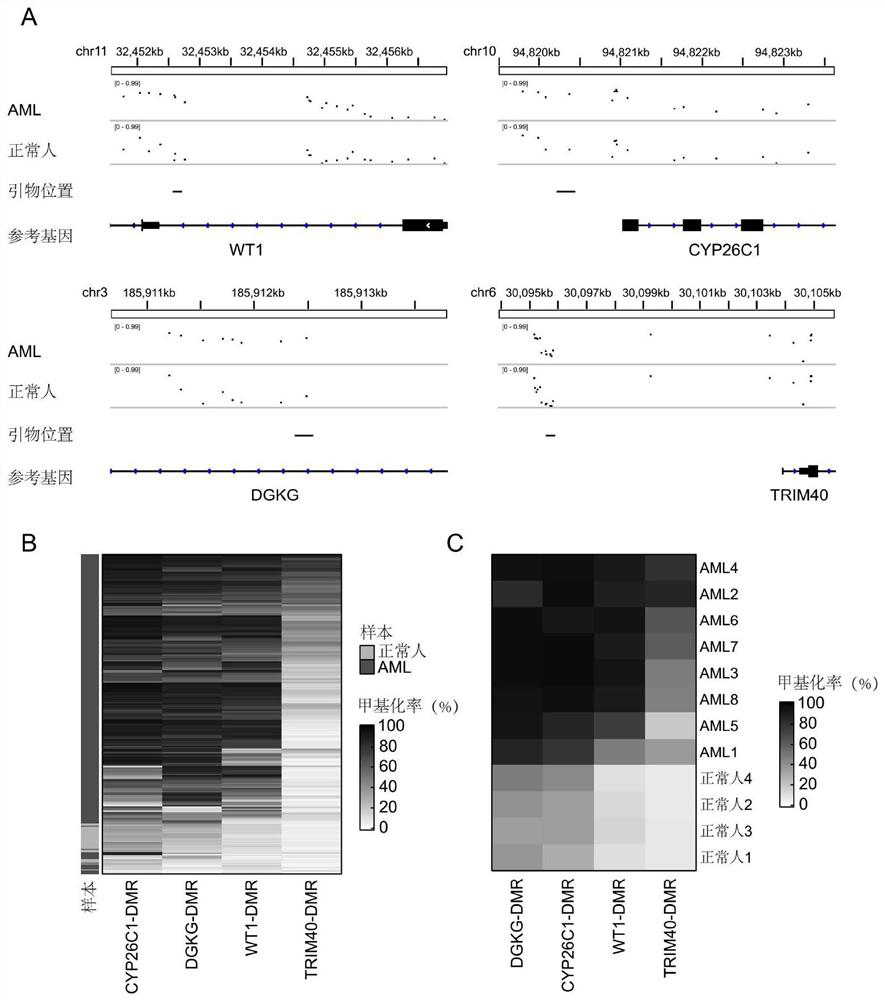
[0026] Example 2 Detection of DNA methylation levels of CYP26C1-DMR, DGKG-DMR, WT1-DMR, TRIM40-DMR and other elements
[0027] Target region DNA methylation sequencing is a method that uses high-throughput sequencing to detect DNA methylation levels in specific regions. It has the advantages of high detection accuracy, parallel detection of multiple sites, and low cost. In order to apply this method to detect the four DNA methylation elements found above, the inventors further narrowed the detection range of the methylation elements, and designed amplification primers for the narrowed methylation element regions (Table 2).
[0028] The specific steps of DNA methylation detection in the target region are as follows:
[0029] 1. Use Novozyme's blood DNA extraction kit (FastPure Blood DNA Isolation MiniKit) to extract peripheral blood DNA. The specific steps are as follows: Take 200 μl of peripheral blood, shake and mix. Add 400 μl of ACK LysisBuffer, mix well, centrifuge at 11...
Embodiment 3
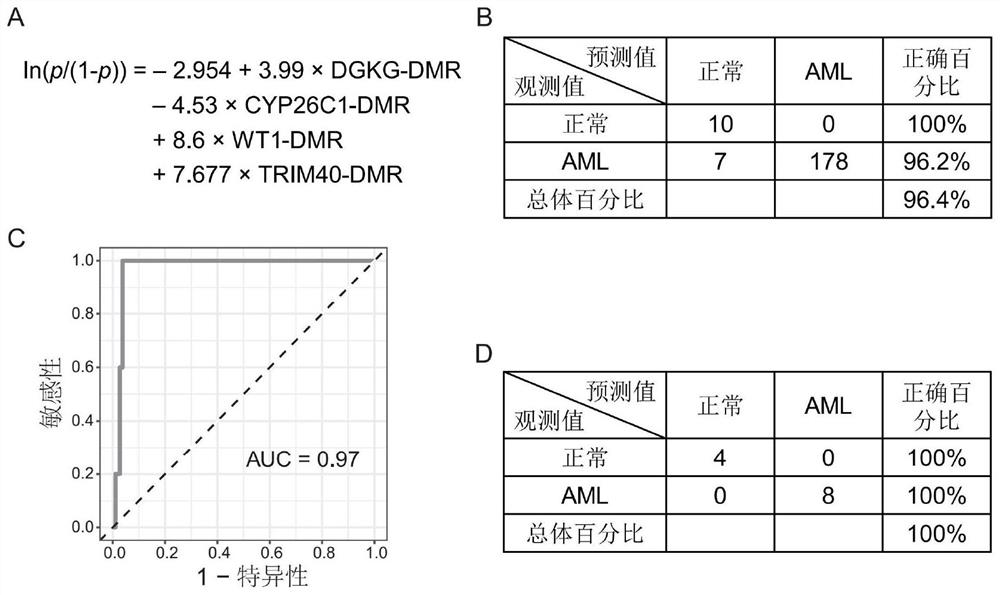
[0035] Embodiment 3 utilizes logistic regression model to diagnose AML
[0036] Using the DNA methylation chip data, the inventors established a logistic regression model for diagnosing AML ( image 3 A), and validate the accuracy of the model using the validation dataset of DNA methylation microarray data. The specific calculation method is: bring the methylation rate of each DNA methylation element into the regression equation, and calculate the p value. If the p value calculated by the equation is greater than 0.5, it is judged as an AML patient, and if it is less than 0.5, it is judged as a normal person. Finally, the model is further validated by using the collected sequencing data of healthy people and AML patients.
[0037] Result: if image 3 As shown in B, the logistic regression model used in the verification data set has a prediction accuracy of 100% for normal people, 96.2% for AML patients, and an overall accuracy of 96.4%. The AUC curve shows that the predicti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com