Microbial synthesis method of eugenol
A microbial synthesis, eugenol technology, applied in the direction of microorganisms, biochemical equipment and methods, botany equipment and methods, etc., can solve the problem of low eugenol content
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
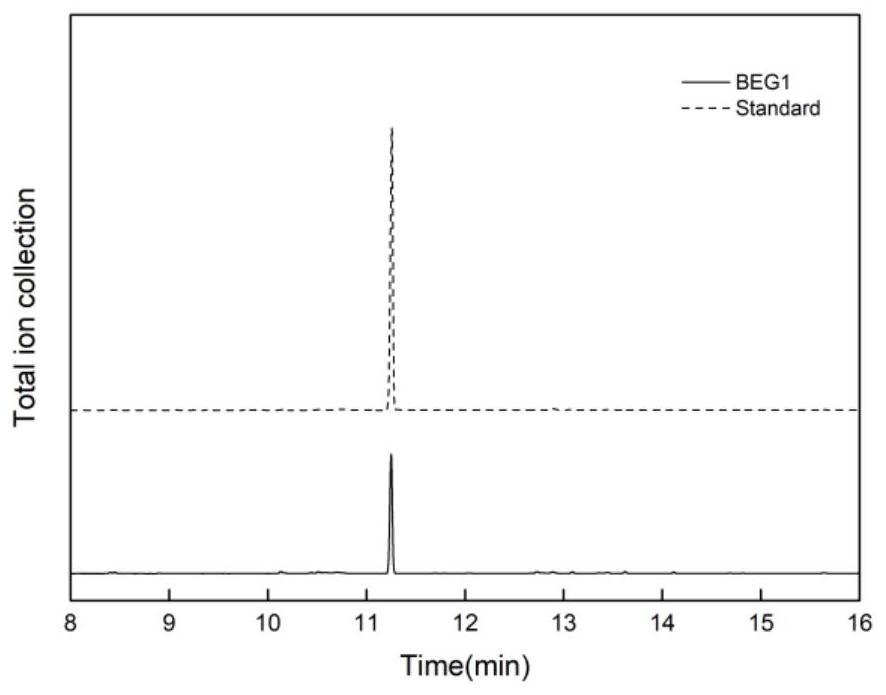
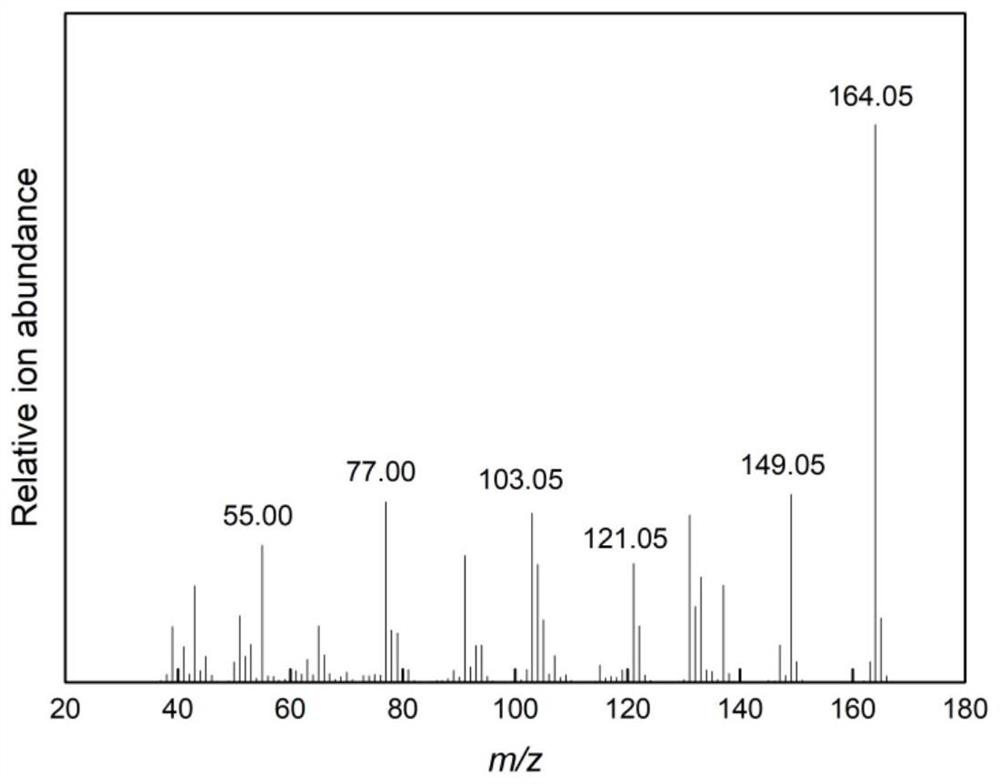
Image
Examples
Embodiment 1
[0031] The acquisition of embodiment 1 (class) coniferyl alcohol acyltransferase
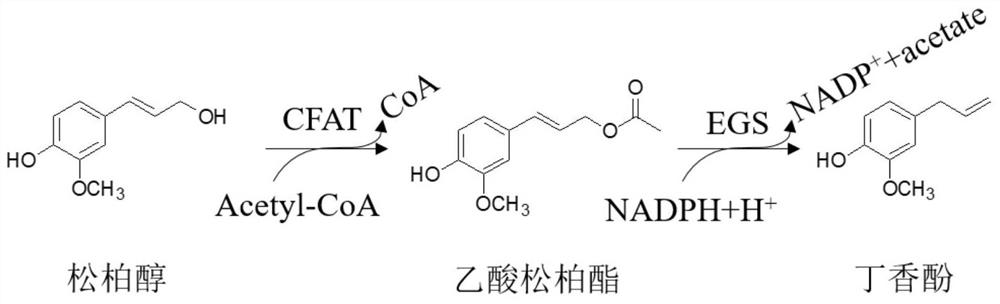
[0032]Coniferyl alcohol acylase can catalyze the acylation reaction of coniferyl alcohol to generate coniferyl acetate. Through bioinformatics analysis and sequence alignment, three coniferyl alcohol acylases were screened out. They are CFAT (GenBank: ABG75942.1 ), CAAT1 (GenBank: KF543260.1) and AT (NCBI: NP_178020.1). The following takes coniferyl alcohol acylase PhCFAT as an example to illustrate how to obtain (like) coniferyl alcohol acylase.
[0033] Combined with the amino acid sequence of coniferyl alcohol acyltransferase CFAT and the codon preference of Escherichia coli, common restriction sites were removed, and a full-length CFAT gene was designed. The nucleic acid sequence is shown in SEQ ID No.2. The CFAT gene was designed to be connected to the downstream of the first T7 promoter of the plasmid pCDFDuet-1, and the P T7 - CFAT gene fragment, the obtained gene fragment is named as s...
Embodiment 2
[0037] The acquisition of embodiment 2 eugenol synthase
[0038] Eugenol synthase catalyzes the reduction reaction of coniferyl acetate to produce eugenol. Four eugenol synthases were screened out through bioinformatics analysis and sequence alignment mining. They are EGS1 (GenBank: ABR24113.1), EGS2 (GenBank: AKB11750.1), APS1 (GenBank: KF543262.1) and AIS1 (GenBank: ACL13526.1), respectively. The following takes eugenol synthase EGS2 as an example to illustrate how to obtain eugenol synthase.
[0039] Combined with the amino acid sequence of eugenol synthase EGS2 and the codon preference of Escherichia coli, the commonly used enzyme cutting sites were removed, and the full-length EGS2 gene was designed. The nucleic acid sequence is shown in SEQ ID No.6. The EGS2 gene was designed to be connected downstream of the second T7 promoter of the plasmid pCDFDuet-1, and the P T7 -EGS2 gene fragment, the obtained gene fragment is named synEGS2.
[0040] The synEGS2 fragment obtai...
Embodiment 3
[0043] Embodiment 3 Construction of recombinant expression vector
[0044] The optimized two types of genes were combined and connected to the plasmid pCDFDuet-1, in which coniferyl alcohol acyltransferase was inserted between the restriction sites Nco I and BamH I, and eugenol synthase was inserted into the restriction site Nde I and Kpn I between. The following takes the recombinant expression vector pCPG1 as an example to illustrate how to construct the recombinant expression vector.
[0045] The synPhCFAT fragment was digested with FastDigest endonuclease Nco I and BamH I, and the reaction system was: 5 μL 10*FD buffer, 2 μL Nco I endonuclease, 2 μL BamH I endonuclease, and 41 μL synCFAT fragment. The reaction conditions are: 37°C, 2h. And use the kit to purify and recover to obtain the synCFAT fragment cut with Nco I and BamH I. The synCFAT fragment obtained after purification and recovery was ligated with the linearized vector backbone pCDFDuet-1 (including the replic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com