Quinoxalinone compounds, compositions, methods, and kits for increasing genome editing efficiency
A compound, alkyl technology, applied to quinoxalinone compounds used to improve genome editing efficiency, can solve problems such as ratios less than 1%
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0691] Example 1: Materials and methods
[0692] method:
[0693] Cells and culture
[0694] Bronchial epithelial cells (BEC) are derived from a human donor diagnosed with cystic fibrosis of the CFTR dF508 / dF508 genotype.
[0695] Induced pluripotent stem cells (iPSC) are derived from human dermal fibroblasts after viral transduction using Yamanaka's reprogramming factors Oct4, Sox2, KLF4 and c-Myc. The derived iPSC can differentiate into 3 germ layers and contains a normal karyotype with 23 pairs of chromosomes.
[0696] Primary human mobilization of peripheral blood (mPB) CD34 + Hematopoietic stem cells and progenitor cells (HSPC) were purchased from Hemacare or AllCells. The cells were thawed, washed and resuspended in a complete medium consisting of serum-free medium CellGro SCGM (CellGenix) and supplemented with a cytokine mixture (300ng / mL SCF, 300ng / mLFlt3L, 100ng / mL TPO, 60ng / mL). ml IL-3), the density is 1-3x 10 5 Cells / mL, and at 37°C / 5% CO before electroporation 2 Incubate...
Embodiment 2
[0730] Example 2: DNA-PK inhibitors improve the ratio of HDR gene editing in BEC
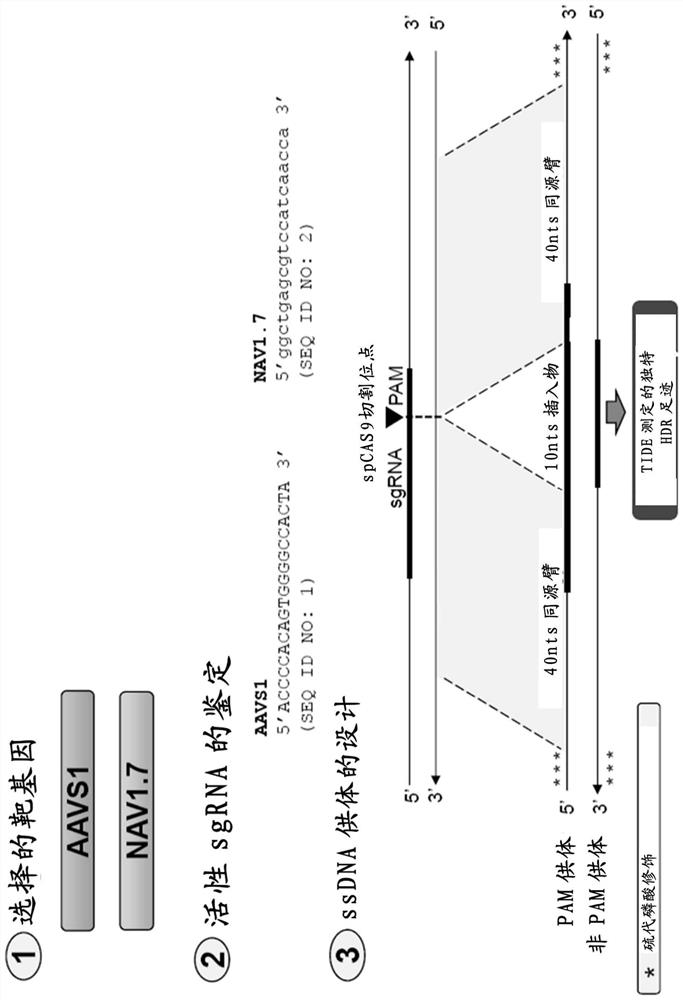
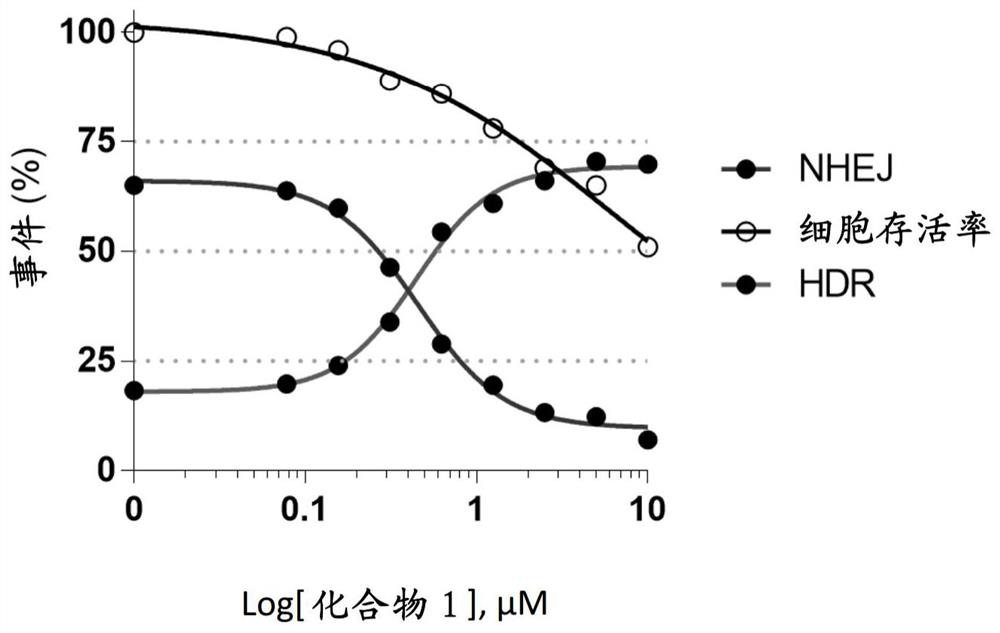
[0731] figure 1 The design of the gene editing assay used in the following examples is exemplified. In order to study the effect of DNA-PK inhibitors on HDR gene editing rate, BEC was electroporated with spCAS9 mRNA, NAV1.7 sgRNA and NAV1.7 non-PAM ssODN, and then incubated with different concentrations of compound 1 or untreated ( Control). The gene editing rate was determined 72 hours after electroporation by using the TIDE assay. The gene editing rate is expressed as a percentage and classified into HDR and NHEJ. The cell survival rate is expressed as a percentage in which the control cell is set to 100%.
[0732] Such as figure 2 As shown in, the DNA-PK inhibitor of compound 1 improves the gene editing rate in BEC. For compound 1, NHEJ IC50 is 0.4450 μM, HDR EC50 is 0.4448 μM, and HDR TOP% is 69.37.
Embodiment 3
[0733] Example 3: DNA-PK inhibitors improve CD34 + HDR gene editing ratio in cells
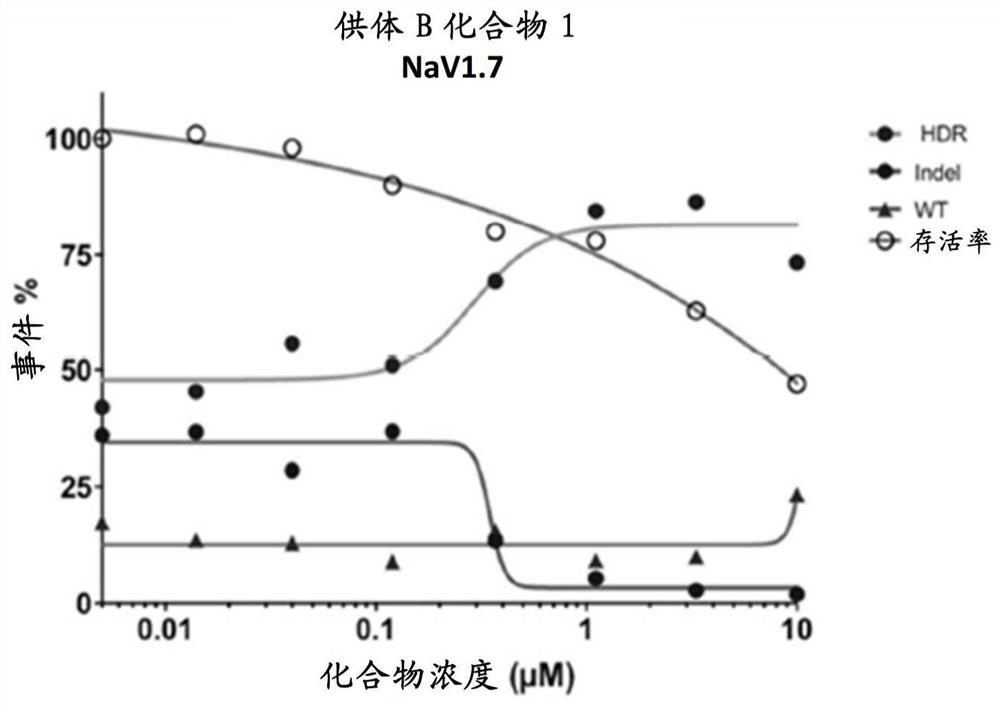
[0734] In order to study the effect of DNA-PK inhibitors on HDR gene editing rate, mPB CD34 was electroporated with RNP (spCAS9 protein + NAV1.7sgRNA) and NAV1.7 non-PAM ssODN + cell. The cells were then incubated with compound 1 at different concentrations. 48h after electroporation, the gene editing rate was determined by using the TIDE assay. The gene editing rate is expressed as a percentage, and such as Figure 3A (Donor B) and Figure 3B (Donor C) is classified as HDR and NHEJ. The cell survival rate was expressed as a percentage in which the control cells were set to 100%.
[0735] Such as Figure 3A with 3B Shown that the DNA-PK inhibitor of compound 1 improves CD34 + The rate of gene editing in the cell. The EC50 values formed by the HDR and Indel of donor B were 0.29 μM and 0.35 μM, respectively.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com