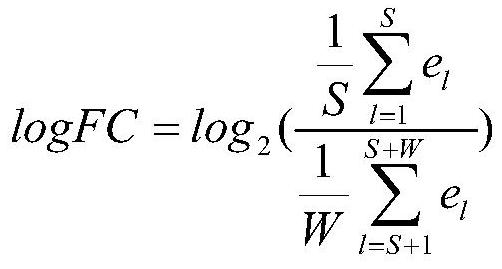Drug relocation method based on differential expression data
A differential expression and gene expression technology, applied in the field of data mining, can solve problems such as low accuracy of results, high false positives, and loss, and achieve the effect of accurate drug prediction accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0034] The embodiments and effects of the present invention will be described in further detail below in conjunction with the accompanying drawings and specific embodiments.
[0035] In this embodiment, breast cancer in the cancer genome TCGA database is taken as an example, and all diseases with pathogenic genes in the TCGA database are used.
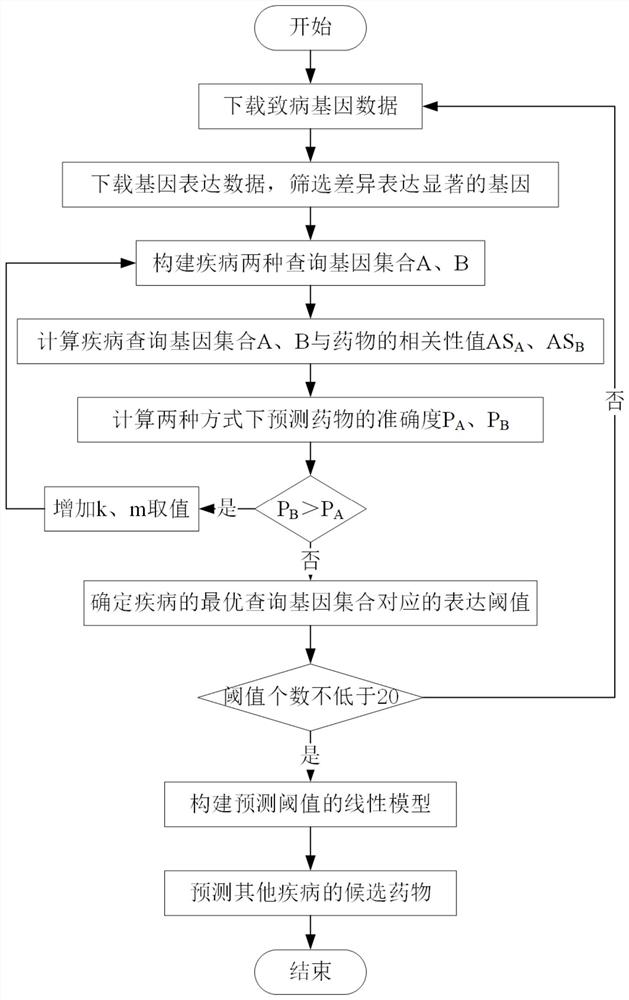
[0036] refer to figure 1 , a drug repositioning method based on differential expression data, the implementation steps are as follows:
[0037] Step 1, download the disease-causing gene data.
[0038] In this example, the OMIM database is used to search for a set of genes causing breast cancer. There are 22 genes in total, which are recorded as set L.
[0039] Step 2, download the gene expression data, calculate the logFC of each gene expression change value and the corresponding false positive FDR, and screen the genes.
[0040] 2.1) Download the breast cancer data and calculate the gene expression change value logFC:
[0041] 2.1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com