3'-terminal-2'-O methylation modified miRNA marker combination related to gastric cancer diagnosis and application thereof
A marker and methylation technology, which is applied in the determination/test of microorganisms, biochemical equipment and methods, etc., can solve the problems of unsatisfactory clinical screening, high cost, and restrict the development of methylation detection, and achieve good application. Prospect, Sensitivity, Specificity, Effectiveness of Broad Health Screening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Screening of 2'-O methylated differential miRNAs at the specific plasma 3' end of patients with gastric cancer
[0040] (1) The subjects of the study were patients with gastric cancer and healthy controls. Plasma from patients with gastric cancer and healthy controls was obtained from Nanjing Eastern Theater General Hospital. The blood collection was approved by patients and healthy volunteers and approved by the Ethics Committee of Eastern Theater General Hospital. The sample consisted of 60 patients with primary gastric cancer and 60 normal healthy volunteers. All patients with gastric cancer were confirmed by biopsy and had not undergone surgery, chemotherapy, radiotherapy or other tumor-related treatments before blood collection. The sample age ranged from 41 to 69 years old, and there was no significant difference in age composition between the two groups (P=0.83). The experiment of the present invention consists of a test set (n=12) and a verification ...
Embodiment 2
[0048] Utilize the kit of the present invention to verify the sequencing results
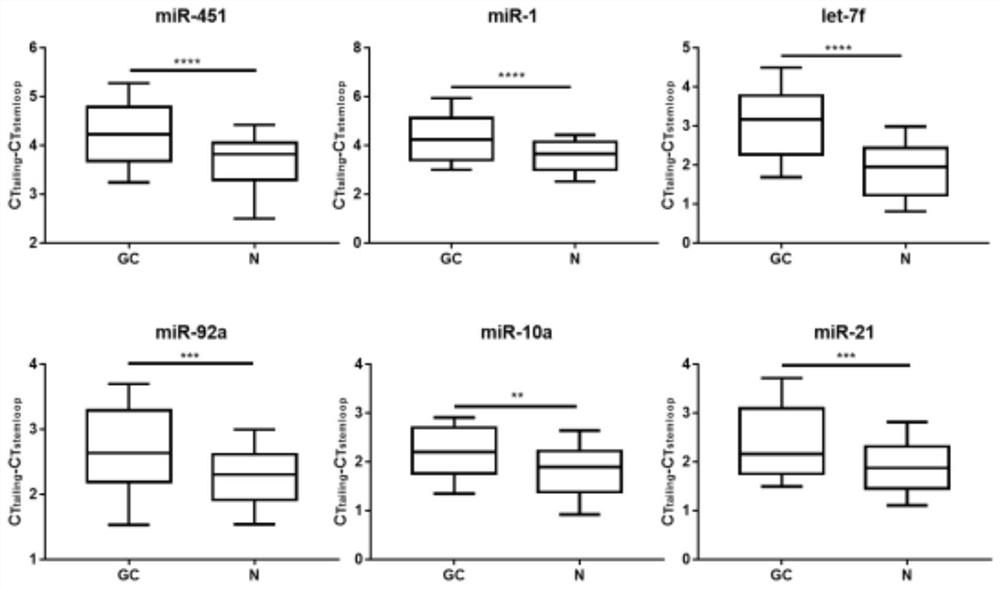
[0049] Stem-loop tailing method to determine the degree of 2'-O methylation at the 3' end of miRNA Principle: The methylation modification at the 3'-end of miRNA can affect the tailing efficiency of the tailing method in the kit due to the existence of steric hindrance , so that the measured miRNA content decreases and the Ct value increases. However, the 2'-O methylation at the 3' end of the miRNA did not affect the miRNA content determined by the stem-loop method, and the Ct value remained unchanged. Therefore, two methods are used to detect the plasma content of miRNA, and the Ct values measured by the two methods are subtracted, and the difference in Ct can reflect the difference in the degree of methylation in plasma miRNA, and explore the use of this method for gastric cancer. Screening diagnosis. The specific test method is:
[0050] (1) Stem-loop method to determine and screen the r...
Embodiment 3
[0071] Operating characteristic curve analysis (ROC curve analysis)
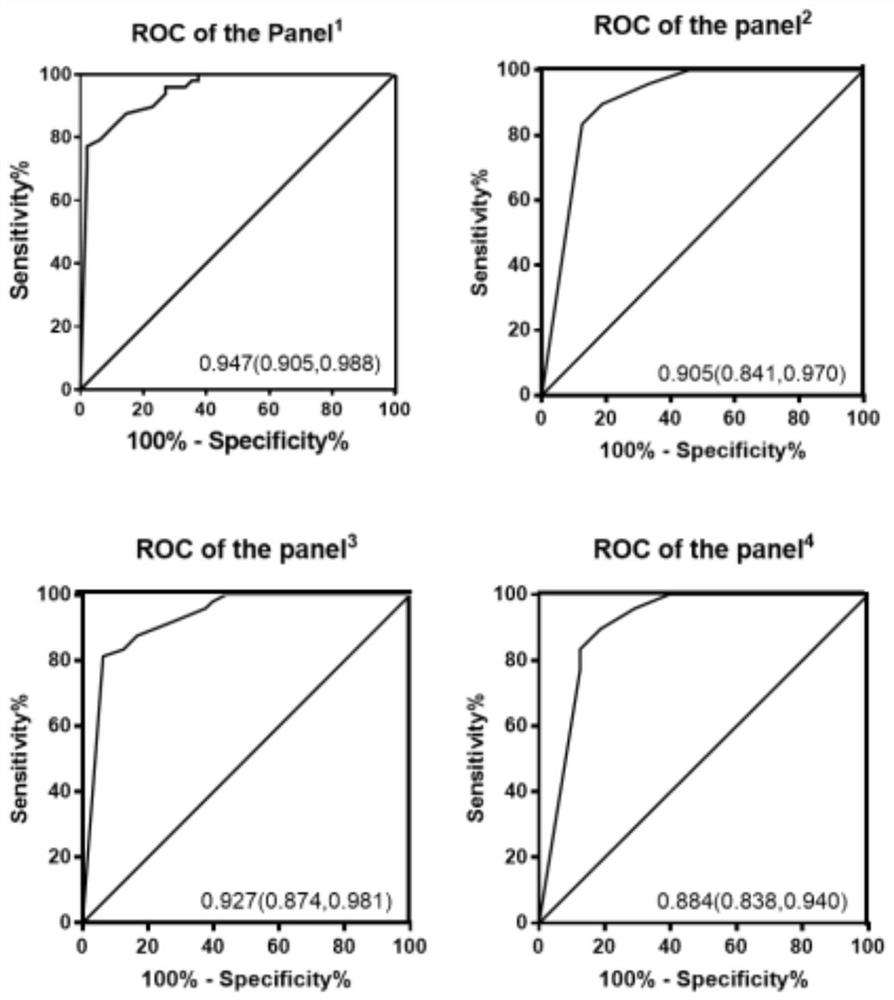
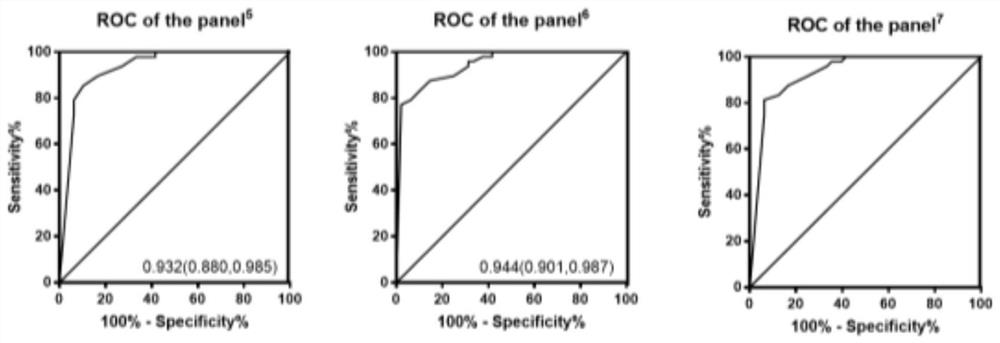
[0072] In order to clarify the diagnostic efficiency of the 6 miRNA methylation assays we screened in patients with gastric cancer, we performed ROC curve analysis on the experimental results. The results are shown in Table 3. The areas under the ROC curve of -2'-O methylation detection to distinguish gastric cancer patients from normal controls were: miR-451, 0.743; miR-1, 0.722; let-7f, 0.840; miR-92a, 0.679; miR- 10a, 0.685; miR-21, 0.691. The area under the curve of the combined detection of 6 miRNAs can reach 0.947, indicating that the combination of 6 miRNAs to detect the 3' end-2'-O methylation level by stem-loop tailing method can significantly screen patients with gastric cancer.
[0073] Table 3: ROC curve analysis of 6 miRNAs.
[0074]
[0075]
[0076] *The panel 1 Represents the joint detection of the above six indicators;
[0077] The panel 2 Represents the combined detection of miR-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com