Genetically engineered bacterium for co-production of 3-hydroxypropionic acid and 1,3-propylene glycol as well as construction method and application of genetically engineered bacterium
A technology of genetically engineered bacteria and hydroxypropionic acid, applied in the field of bioengineering, can solve the problems of affecting the reaction, unbalanced coenzymes, low yield, etc., and achieve the effects of efficient metabolism, increased yield, and removal of cytotoxicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
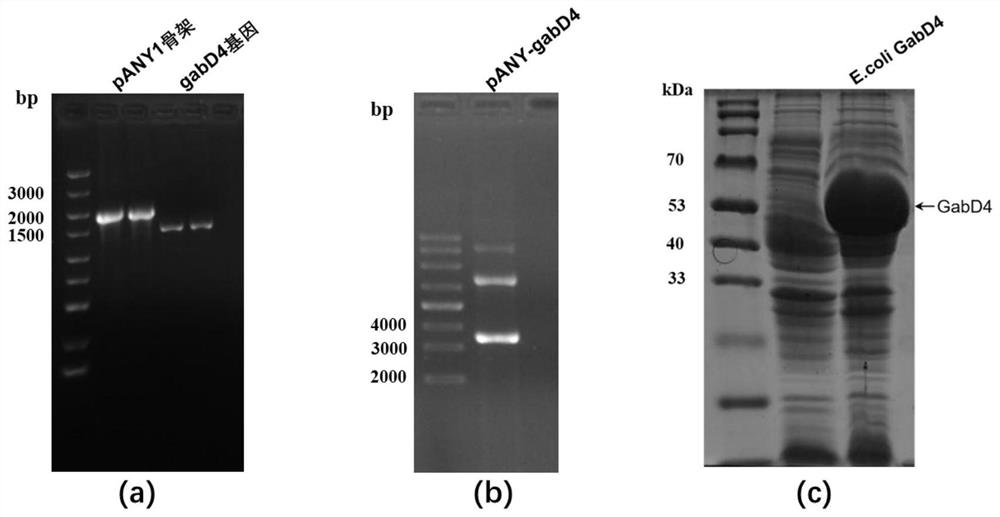
[0031] Example 1: Recombination E. coli Construction and expression of BL21 / pANY-GabD4
[0032] (1) According to the gene encoding succinic semialdehyde dehydrogenase of Copper-loving hookworm GabD4 Sequence and characteristics of gene elements on the vector pANY1, using Oligo7.0 software to design primers: PANY-F: 5'-atgtatatctccttcttaaagt-3', PANY-R: 5'-cctccatgggagctcctg-3', GabD4-F1: 5'-taactttaagaaggagatatacatatgtaccaagatctggcactgt- 3' and GabD4-R1: 5'-tgcaggagctcccatggaggttacgcttgggtgatgaact-3'.
[0033] (2) Use PANY-F and PANY-R primer pairs to amplify the pANY1 vector backbone (excluding 6×His tag and ccdB expression cassette), PCR reaction parameters: pre-denaturation, 98°C 1min; denaturation, 98°C 10s; annealing, 55°C for 10s; extension, 72°C for 30s; stop extension, 72°C for 5min; pANY1 vector backbone was obtained after 32 cycles.
[0034] GabD4-F1 and GabD4-R1 primer pairs were used to amplify homology-arm containing GabD4 Gene fragments, PCR reaction par...
Embodiment 2
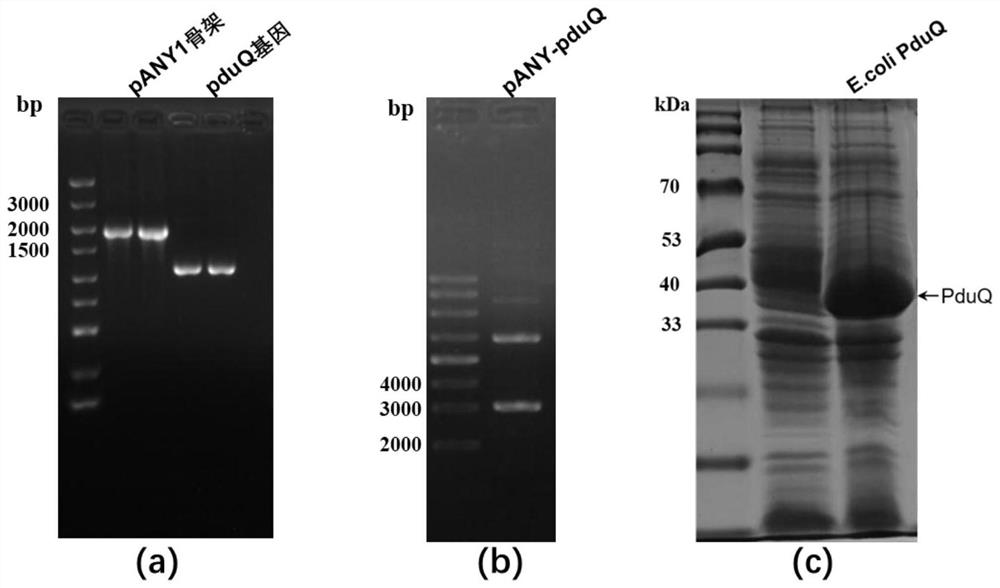
[0039] Example 2: Construction and expression of recombinant E.coli BL21 / pANY-PduQ
[0040] (1) According to the gene pduQ sequence of Lactobacillus reuteri encoding 1,3-propanediol oxidoreductase and the characteristics of the gene elements on the vector pANY1, use Oligo7.0 software to design primers: PduQ-F1: 5'- taactttaagaaggagatatacatatggaaaaatttagtatgccaac-3' and PduQ-R1: 5'-tgcaggagctcccatggaggttaacgaattattgcttcgtaaat-3'.
[0041] (2) Use PANY-F and PANY-R primer pairs to amplify the pANY1 vector backbone (excluding 6×His tag and ccdB expression cassette), PCR reaction parameters: pre-denaturation, 98°C for 1 min; denaturation, 98°C for 10s; annealing , 55°C for 10s; extension, 72°C for 30s; stop extension, 72°C for 5min; pANY1 vector backbone was obtained after 32 cycles.
[0042] Use the PduQ-F1 and PduQ-R1 primer pairs to amplify the pduQ gene fragment containing the homology arm, PCR reaction parameters: PCR reaction parameters: pre-denaturation, 98°C 1min; denatu...
Embodiment 3
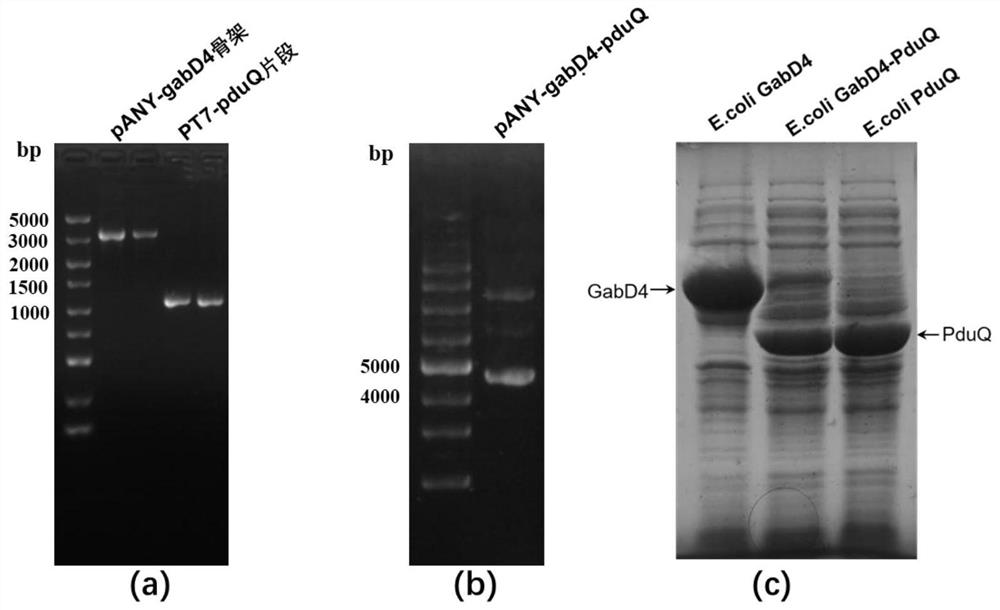
[0047] Example 3: Construction and expression of recombinant E.coli BL21 / pANY-GabD4-PduQ
[0048] (1) Use Oligo7.0 software to design primers according to the sequences of vectors pANY-gabD4 and pANY-pduQ successfully constructed in Examples 1 and 2: GabD4-F2: 5'- cctccatgggagctcctg-3', GabD4-R2: 5'- ttacgcttgggtgatgaactt-3', PduQ-F2: 5'-agttcatcacccaagcgtaatggccttttgctgg-3' and PduQ-R2: 5'-tgcaggagctcccatggaggttaacgaattattgcttc-3'.
[0049] (2) E. coli GabD4 and E. coli PduQ plasmids pANY-gabD4 and pANY-pduQ were extracted using the plasmid mini-extraction kit, respectively. Using the plasmid pANY-gabD4 as a template, use the primer pair GabD4-F2 and GabD4-R2 to linearize pANY-gabD4. PCR reaction parameters: pre-denaturation, 98°C 1min; denaturation, 98°C 10s; annealing, 55°C 10s; extension , 72°C for 30s; stop extension, 72°C for 5min; pANY-gabD4 backbone was obtained after 32 cycles.
[0050] Using the plasmid pANY-pduQ as a template, use PduQ-F2 and PduQ-R2 primers to am...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap