Anti-COVID-19 drug discovery method based on network characterization
A discovery method and characterization technology, applied in the field of computer applications and bioinformatics, can solve problems such as limited understanding, and achieve the effect of improving performance, improving prediction accuracy, and improving performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0039] The present invention will be further described below in conjunction with the accompanying drawings.
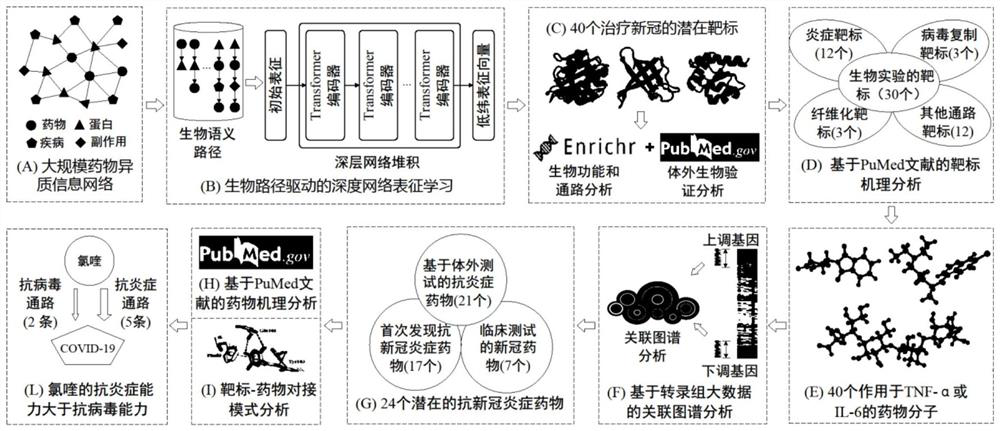
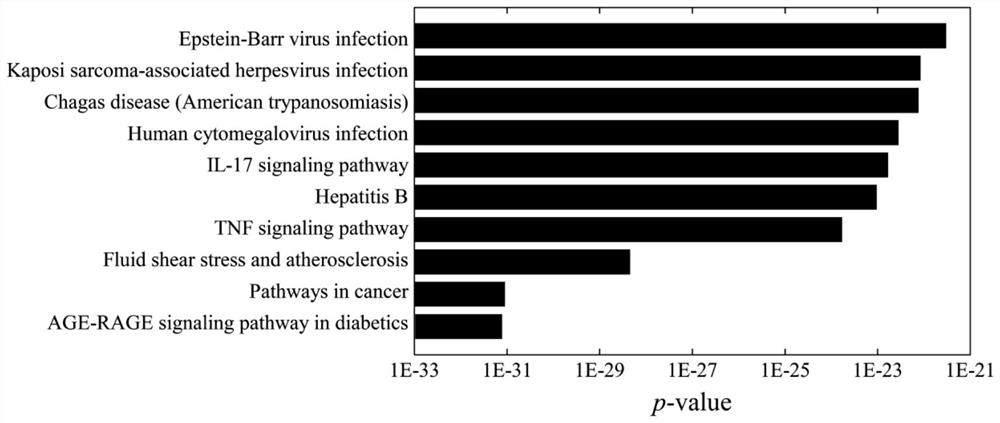
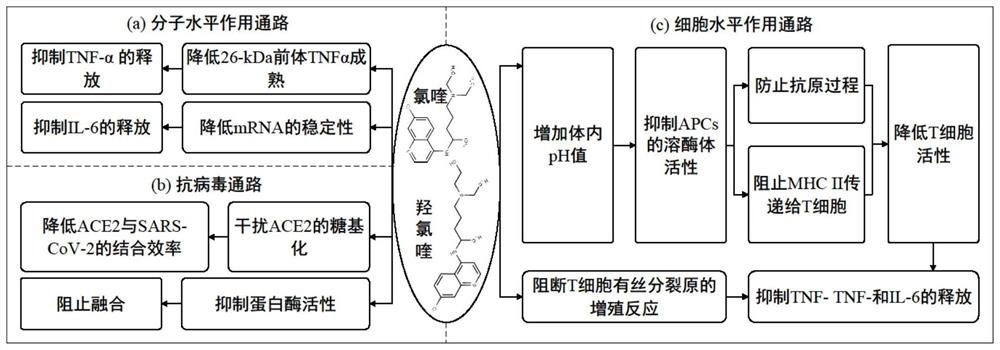
[0040] refer to figure 1 , a method for discovering anti-coronavirus inflammatory drugs based on network characterization, the method comprising the following steps:
[0041] 1) Parameter initialization, including setting the number of sequence trajectories psize, network sequence length l, node reading threshold deg, representation vector dimension dim, Transformer encoder layer number n;
[0042] 2) Build a drug heterogeneous information network;
[0043] 3) Randomly select psize∈[1,num] and psize∈N + nodes as the initial sampling node x of each sequence trajectory j ∈{x i |i=1,2,...,num}, sequentially sample the network node sequence according to the specific semantic path;
[0044] 4) Perform word segmentation on all sampled sequences, including unicode string conversion, remove special characters, space word segmentation, remove redundant characters and punct...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com