Clustering method of gene microarray containing missing values based on joint training
A technology of gene microarray and clustering method, which is applied in the field of clustering of gene microarrays with missing values
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
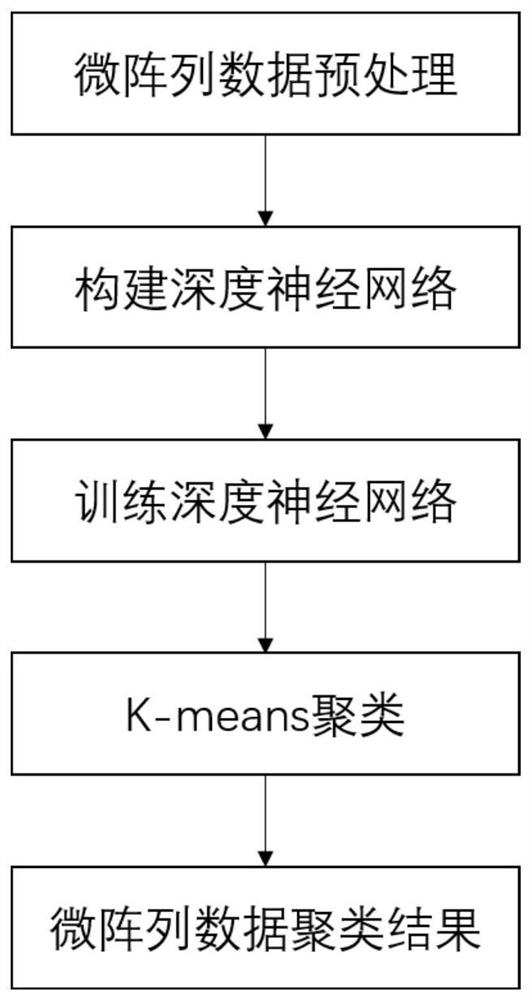
[0042] Such as figure 1 As shown, this embodiment discloses a clustering method for gene microarrays containing missing values based on joint training. First, a sample of the gene microarray data set is obtained, and the sample of the gene microarray data set is a data matrix, and the gene microarray data Divided into a training set and a test set, in the present embodiment, the data matrix is composed of 500 row vectors, each row vector of the data matrix corresponds to the expression data sequence of the gene, and 500 represents the number of genes recorded in the data matrix; then, construct A deep neural network based on joint training of gene microarrays containing missing values, wherein the deep neural network includes a Sequence-To-Sequence encoding-decoding network and an adversarial learning module; use the training set to train the deep neural network and determine the deep neural network The learnable parameters in the test set are input into the deep neural ne...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com