Fluorescent quantitative reference genes of different tissue of cryptomeria fortunei as well as special primer and application of fluorescent quantitative reference genes
A fluorescence quantitative, internal reference gene technology, applied in the field of plant molecular biology, to achieve the effects of high amplification efficiency, improved detection efficiency, and improved reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] The screening of the most stably expressed internal reference gene in different tissues of Cryptomeria comprises the following steps:
[0031] 1. Sampling of test materials: Collect cedars that grow well in the Baima base of Nanjing Forestry University, and collect cedar roots, stems, leaves and fruits in July 2020, store them in dry ice and bring them back to the laboratory, and place them in a -80°C refrigerator save.
[0032] 2. Extraction of total RNA from different tissues: The total RNA from different tissues of Cedar cedar was extracted by using the general-purpose total RNA extraction kit of Biotech Biotechnology Company according to the instructions.
[0033] 3. Synthesis of the first strand of cDNA:
[0034] The cDNA reverse transcription kit ( III RT SuperMix for qPCR (+gDNA wiper) R323-01) reverses the total RNA to eDNA as a template for fluorescent quantitative PCR.
[0035] 1) Prepare the following mixture in an RNase-free centrifuge tube: 4x gDNA wipe...
Embodiment 2
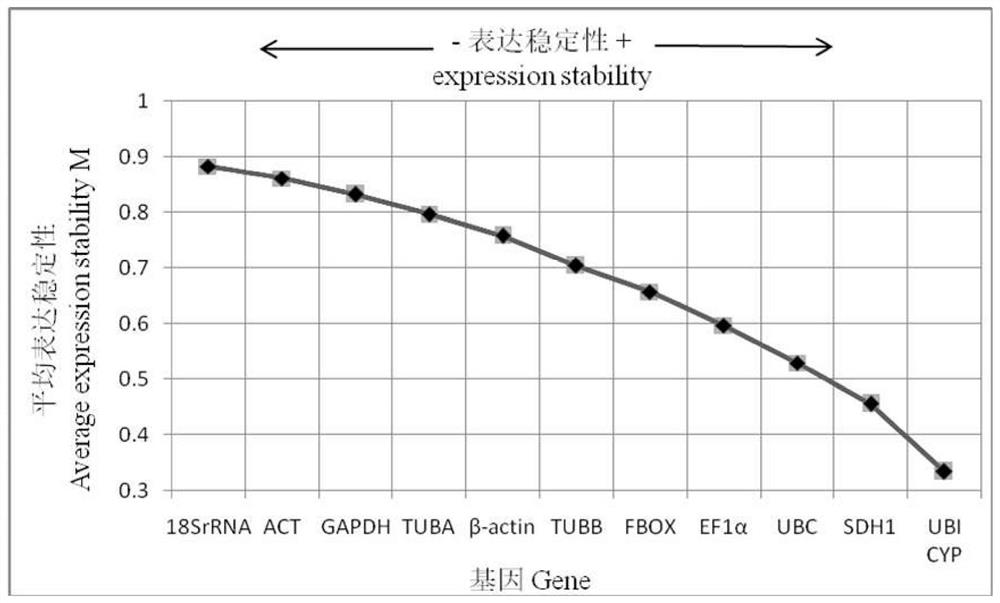
[0063] Three internal reference genes UBI, CYP, and SDH1 with comprehensive ranking stability and unstable internal reference gene 18S rRNA were used for verification.
[0064] In order to verify the stability of the internal reference genes screened, the expression patterns of CESA, CHS and PYL in different tissues of Cryptomeria were analyzed.
[0065] The specific primers for the CESA gene are:
[0066] CESA-F: 5'-CCAACGGTCAAGTGGTGTCT-3';
[0067] CESA-R: 5'-GGTTAGGGTCAGCATAGGGA-3'.
[0068] The specific primers for the CHS gene are:
[0069] CHS-F: 5'-ATGCCAAATGTCGCCAAAGT-3';
[0070] CHS-R: 5'-GTCGTTTCTCAAGAGCGTGCC-3'.
[0071] The specific primers for the PYL gene are:
[0072] PYL-F: 5'-CACAGGCTACGGGATTACCA-3':
[0073] PYL-R: 5'-GCAAATTACACCGCACAACG-3'.
[0074] In the expression of CESA, CHS and PYL ( Figure 4 , Figure 5 and Figure 6 ), when the unstable internal reference gene 18S rRNA was used as the internal reference gene, it was highly expressed in s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com