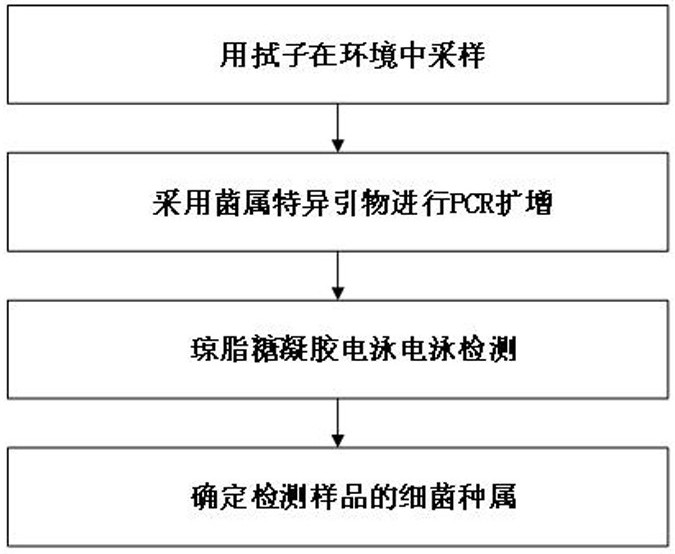
A primer set, kit and method for identifying environmental bacteria at the genus level
An environmental and bacterial technology, applied in biochemical equipment and methods, microbial measurement/inspection, recombinant DNA technology, etc., can solve the problems of inability to detect bacteria extensively and comprehensively, and achieve low cost, short time consumption, and simple operation Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Species Identification and Classification of Environmental Bacteria
[0063] Specific steps are as follows:
[0064] A. Use swabs to sample bacteria from different locations in the environment (experimental benches, instrument surfaces, etc.) (sampling liquid is bacterial lysate), numbered 1~48 in sequence.
[0065] B. Gradiently dilute the sample solution to 10 -6 , take 10 respectively -4 , 10 -5 , 10 -6 10 μl of bacterial solutions of three concentrations were spread on LB solid medium, and cultured overnight in a 37°C incubator. After colonies grew, single colonies were picked and streaked three times for isolation and culture to obtain pure bacteria.
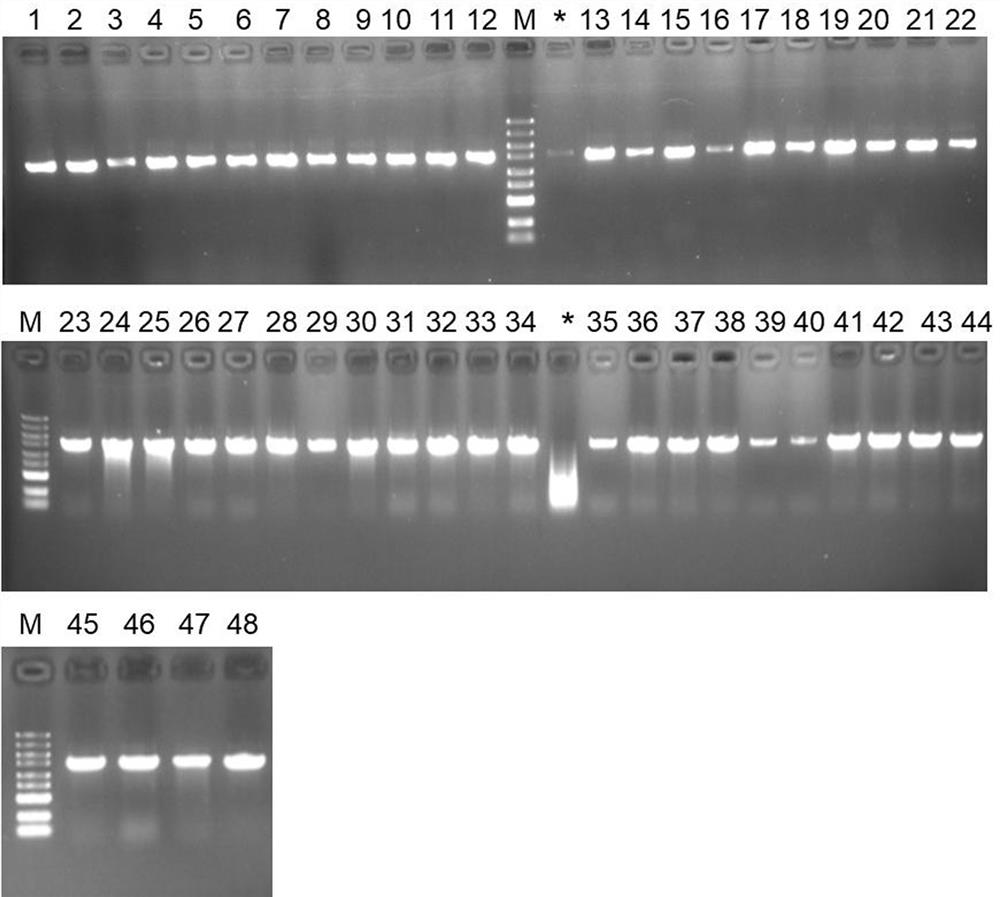
[0066] C. Pick the single bacterial colony of the purified streak culture, carry out PCR amplification with 16S rDNA universal primers 27F and 1492R, and detect the PCR amplification results by agarose gel electrophoresis (see the electrophoresis results in figure 2 , where 1~48 are 48 collected samples, M is M...
Embodiment 2
[0083] Design and detection of genus-level specific primers
[0084] Specific steps are as follows:
[0085] A, according to the sequencing results of 48 kinds of bacteria in Example 1, and search for differential sequences, design 16S rDNA bacteria-specific primers, wherein R1 is a specific reverse primer for Pantoea, and R2 is Staphylococcus, Bacillus, Micrococcus, Pseudomonas A specific reverse primer common to both Monas and Bacillus.
[0086] Staphylococcus specific forward primer F1: 5'-ATGTGTAAGTAACTATGCACGTCT-3' (SEQ ID No.1)
[0087] Bacillus-specific forward primer F2 5'-CTAGTTGAATAAGCTGGCACCT-3' (SEQ ID No.2)
[0088] Pantoea specific forward primer F3 5'-CAGCGATTCGGTCGGGAACTCA-3' (SEQ ID No.3)
[0089] Micrococcus specific bacteria forward primer F4 5'-AGCGAAAGTGACGGTACCTGCAGAAG-3' (SEQ IDNo.4)
[0090] Pseudomonas specific forward primer F5 5'-GCAGTAAGTTAATACCTTGCTGTTT-3' (SEQ ID No.5)
[0091] Aerobicella specific forward primer F6 5'-ATTGTAGAGTAACTGCTACAGTCT-...
Embodiment 3
[0114] Identifying Environmental Bacteria at the Genus Level
[0115] Specific steps are as follows:
[0116] A. Use a swab to sample bacteria from different locations in the environment (1, table 2, window 3, glass 4, small trash can 5, doorknob 6, floor) (the sampling liquid is lysate).
[0117] B. Using the collected sample as a template, carry out PCR amplification with the forward primer and reverse primer in Example 2, respectively.
[0118] C. Perform agarose gel electrophoresis on the PCR product.
[0119] D. Results of agarose gel electrophoresis ( Figure 5 ) for analysis. From Figure 5 It can be seen that the provided primer pairs are specific and can be used to directly amplify bacterial templates sampled at different locations in the environment, and the bacterial species contained in different locations in the environment are different.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com