A method for predicting lncRNA-mirna interactions in plants
A plant and prediction matrix technology, which is applied in the field of plant biomolecular relationship prediction, can solve the problems that lncRNA or miRNA similarity prediction cannot be applied, and achieve the effect of speeding up and enhancing accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0059] In order to make the object, technical solution and advantages of the present invention clearer, the present invention will be further described in detail below in conjunction with the accompanying drawings and embodiments. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention. In addition, the technical features involved in the various embodiments of the present invention described below can be combined with each other as long as they do not constitute a conflict with each other.
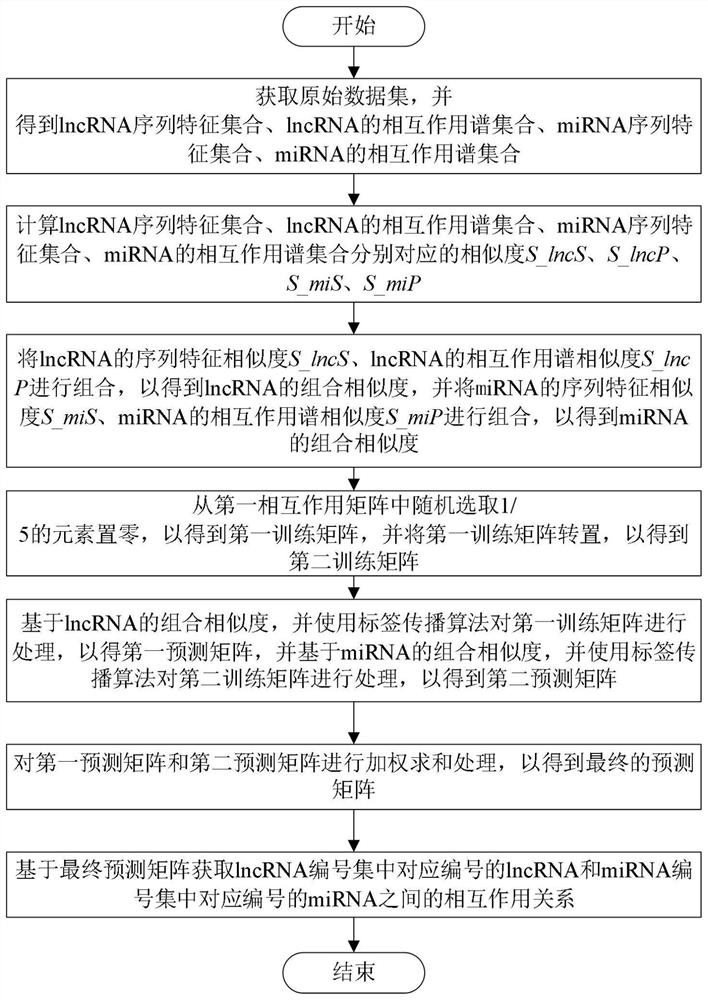
[0060] like figure 1 As shown, the present invention provides a method for predicting plant lncRNA-miRNA interaction, comprising steps:
[0061] (1) Obtain the original data set, and process it to obtain the lncRNA sequence feature set, the lncRNA interaction profile set, the miRNA sequence feature set, and the miRNA interaction profile set;
[0062] The advantage of this step is that the requ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com