SNP primer set and application for individual identification and paternity test of giant panda
A paternity test, giant panda technology, applied in the biological field, to achieve the effect of perfect genealogy, accurate paternity test and individual identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1 Preparation of SNP primer sets for individual identification and paternity identification of giant pandas
[0020] (1) Extract the DNA of 56 giant pandas from 7 groups of families (at least 3 individuals in each group);
[0021] Shred approximately 500 uL of blood clot in a 2ml EP tube with sterilized scissors; add an equal volume of lysis buffer in sequence, containing TNE (1X) and 20% volume of SDS (10%) in the lysis buffer Mix well with 30uL proteinase K (20mg / ml), shake in a water bath, and digest overnight at 55°C; cool the solution to room temperature, add an equal volume of Tris-saturated phenol, shake in a constant temperature shaking box for 8-12h, Centrifuge at 12000rpm for 10 minutes, transfer the supernatant to another centrifuge tube; add an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1) mixture, shake in the shaking box for 2-3 hours, then take out the centrifuge at 12000r / min After 10 minutes, take the supernatant; add an equal v...
Embodiment 2
[0035] Example 2 Detection of SNPs with individual resequencing methods
[0036] (1) Use the BWA comparison software to compare the clean data of each sample to the panda genome (ftp: / / ftp.ncbi.nlm.nih.gov / genomes / all / GCA / 000 / 004 / 335 / GCA_000004335 .1_AilMel_1.0) to get the initial alignment result file in BAM format. Use the Picard tool to remove duplicate reads (the duplication here refers to the identical reads caused by PCR amplification) and sort according to the alignment position.
[0037] (2) Use the Haplotype Caller tool of GATK software to simultaneously detect SNPs and Indels (the alignment file *.BAM used for detection has been corrected for base quality values and incorrect alignment positions caused by indels).
[0038] (3) The SNP and Indel marked as "PASS" in the output VCF result file are credible variation sets that have passed the filtering conditions.
Embodiment 3
[0039] Example 3 Paternity Test / Individual Identification SNP Screening:
[0040] (1) For individual samples with parent-child / mother-child relationship, retain SNPs with an allele frequency greater than 0.4 and no Indel within the flanking 500bp range;
[0041] (2) According to the pedigree chart, find 7 groups of families with obvious genetic relationship (at least 3 individuals in each group), and screen the SNPs that stably exist in each family;
[0042] (3) Retain SNPs that exist stably in each family with a distance greater than 10Mb.
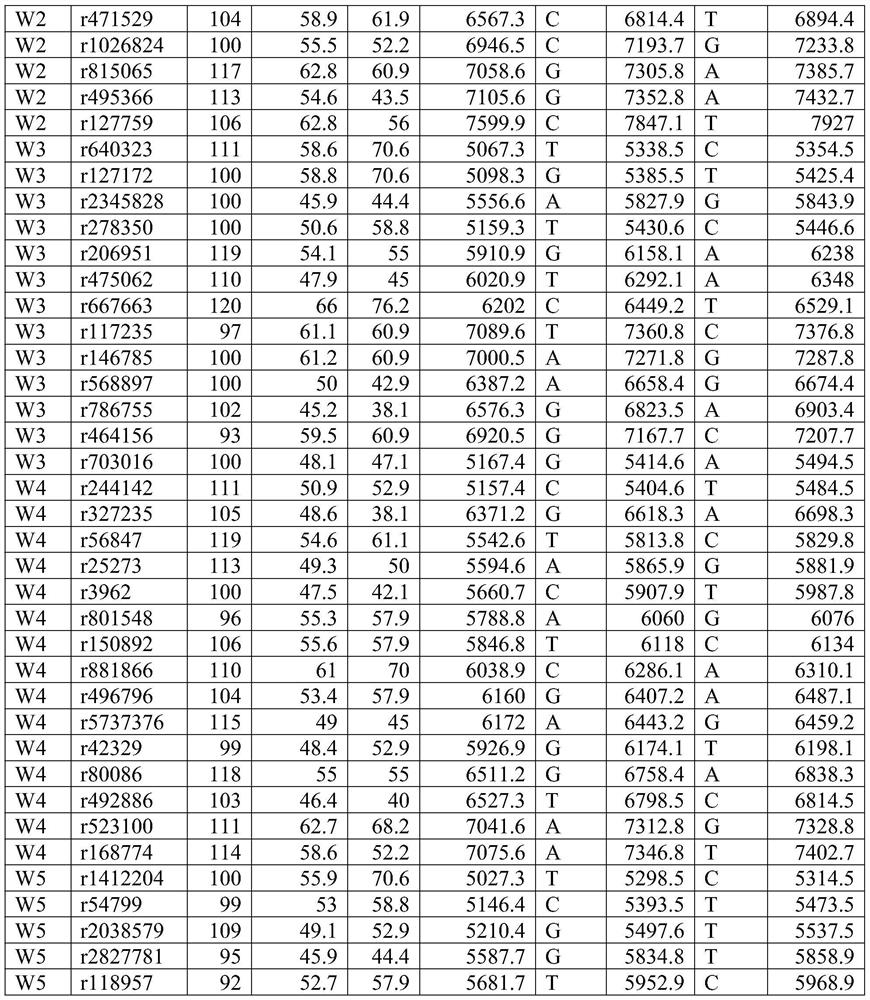
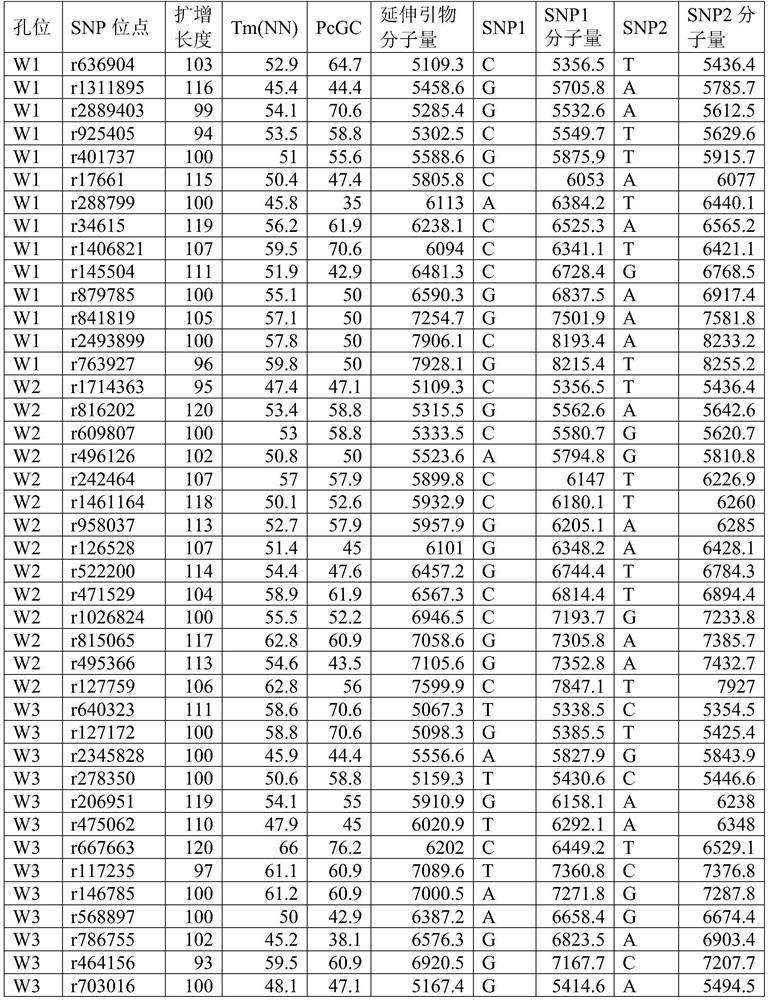
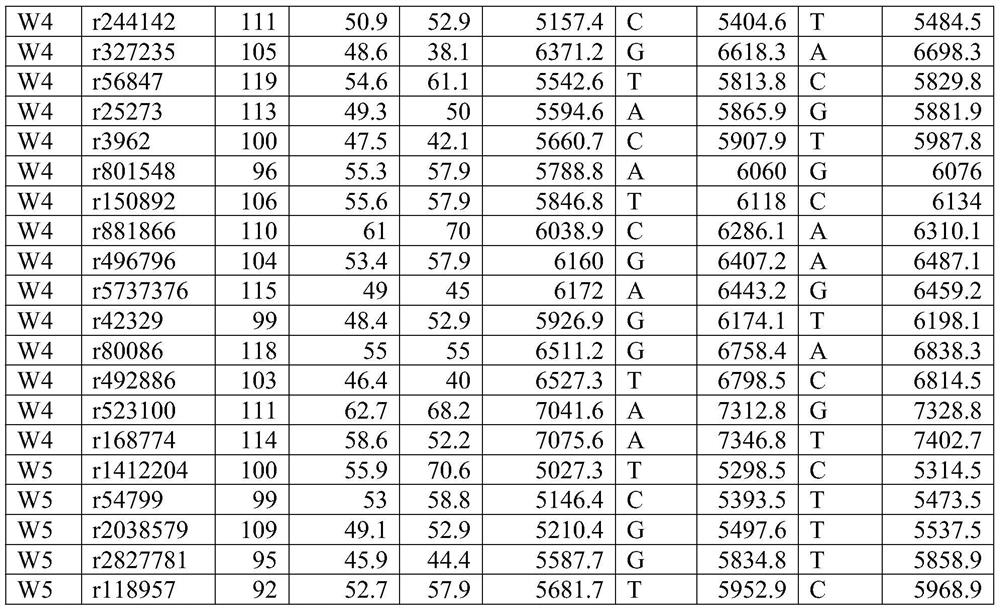
[0043] According to the above requirements, 514 SNP markers that can be used for paternity identification and individual identification were screened from the genome.
[0044] Primer design:
[0045] Primers were designed with Primer 5.0. Among them, the length of the amplified fragment is controlled between 45-120bp; primer mismatch and hairpin structure are avoided; the TM value is controlled between 45°C and 65°C. The 3' end of the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com