Compositions and methods for targeting mutant ras
A technology of mutant and composition, applied in the field of compositions and methods for targeting mutant RAS, can solve the problem of no RAS oncoprotein and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0539] Example 1: Identification of mRAS neoantigens
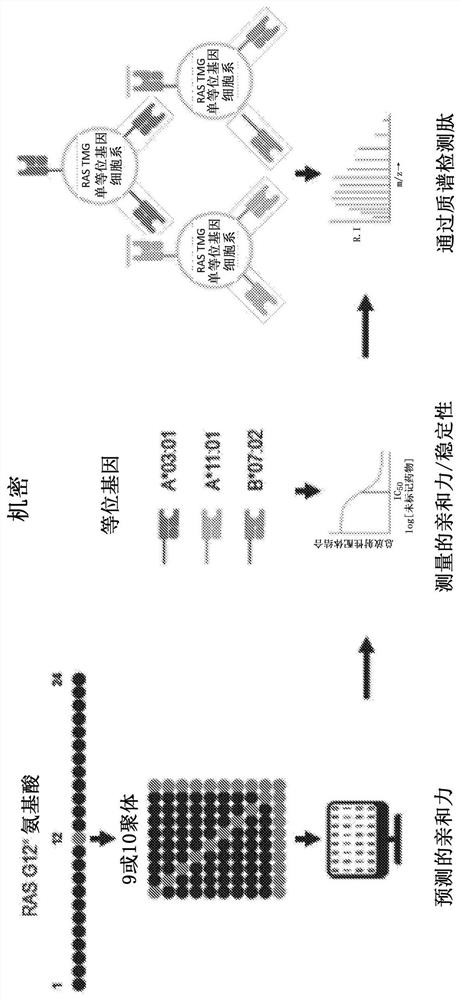
[0540] As described here, various computational and proteomic studies were performed to identify mRAS neoantigens and their interactions with HLA types. figure 1 is a schematic depicting a discovery strategy for mutant RAS epitopes, in which a computer model is used to predict the affinity of mRAS peptides for MHC, followed by experiments to measure the affinity / stability of the interaction and detect the peptides by mass spectrometry.
[0541] A computer study was conducted to predict mRAS neoantigens using antigen.garnish software, which analyzes human or mouse DNA missense mutations, insertions, deletions and fusions and predicts neoepitopes computationally using 7 efficient algorithms . The model exports neoepitopes by MHC I / II binding affinity. For example, if figure 2 As shown, the model was used to predict a 9-10-mer neoepitope containing a mutation at a position corresponding to G12 in RAS.
[0542] Such as ...
Embodiment 2
[0548] Example 2: Evaluation of Immunogenicity of mRAS
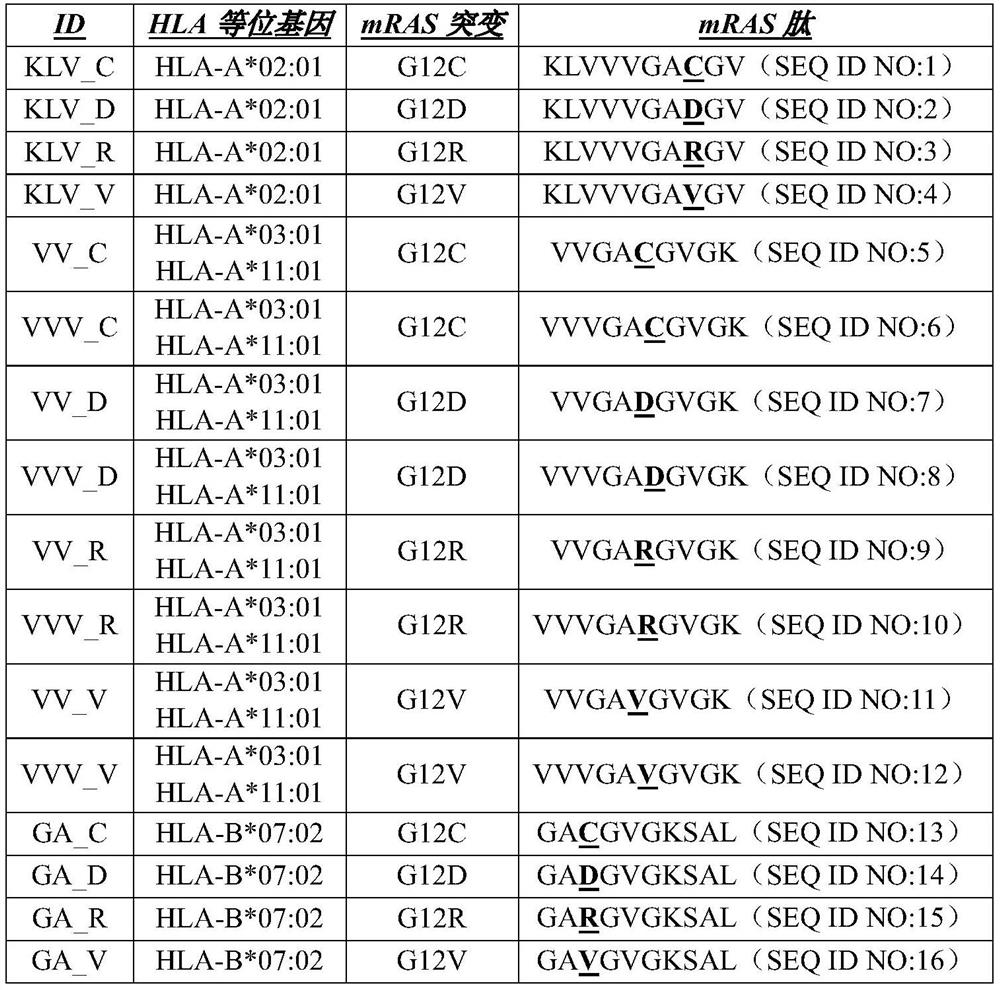
[0549] Experiments were performed to assess the immunogenicity of mRAS peptide-MHC recognition as described herein. Such as Figure 11 As indicated, PMBCs were isolated and stimulated from normal donors selected for HLA type (HLA-A02, HLA-A03, HLA-A11 and HLA-B07). Experiments using the IFN-γ ELISPOT assay were performed to assess CD8+ T cell responses. Summary of mRAS CTL responses as Figure 12 as well as Figure 13A-Figure 13F shown.
[0550] Additional experiments using pMHC multimer staining were performed on T cell cultures to identify mRAS-specific T cells ( Figure 14 ). Experiments also showed that mRAS T cell responses were highly specific ( Figure 15 ). Experiments using various doses of B7-G12R were used to demonstrate the observed response with high affinity ( Figure 16 ).
[0551] Experiments were also performed to examine whether the B7-G12R CTL response could kill the G12R+PDA cell line. Su...
Embodiment 3
[0553] Example 3: Development of mRAS-specific TCR therapy
[0554] As described herein, experiments were performed to design TCR therapies targeting specific mRAS peptides in the context of specific HLA types.
[0555] Such as Figure 18 As indicated, T cells observed to bind A11-G12V and B7-G12R mRAS peptide-HLA type complexes were sorted, expanded, and subjected to TCRα / β sequencing. Two distinct CD8+ T cell clones were identified as HLA-A*11:01-restricted G12V-specific T cells: (1) TRAV39 / TRBV20-1 and TRAV12-1 / TRBV28 were observed.
[0556] Based on the sequencing, lentiviral constructs called TCR831 and TCR833 were designed ( Figure 19 ). TCR831 contains TRAV39 and TRBV20-1, while TCR833 contains TRAV12-1 and TRBV28. Both constructs contain the T2A linker domain and the EF1α promoter. Similarly, additional constructs were designed, termed TCR896, TCR897, TCR847 and TCR864, as Figure 20 summarized. Figure 20 RAS mutation sensitivity, HLA restriction, α-chain an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com