Detecting cancer, cancer tissue of origin, and/or cancer cell type
A cancer, type of technology, applied in the detection of cancer, cancer-derived tissue and/or cancer cell types, can solve problems such as difference, loss of confidence in small control groups, and difficulty in interpretation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0297] Example 1: Analysis of Probe Quantities
[0298] To test how much overlap is needed between a cfDNA fragment and a probe to achieve a non-negligible amount of pulldown, various lengths of overlap were used designed to include three different types of probes (VID3, VID4, VIE2). Detection combinations are tested. The three different types of probes had various overlaps with several 175 bp target DNA fragments specific for each probe. The overlap range tested was between 0bp and 120bp. Several samples including 175bp target DNA fragments were applied to the detection assembly and washed, and then several DNA fragments linked to the several probes were collected. The amount of several DNA fragments collected was measured, and the amount was plotted as density versus size of the overlap, as in Figure 10 provided in .
[0299] When less than 45 bp overlap, there is no significant binding and pull-down of the target DNA fragment. These results show that a fragment-probe ...
example 2
[0303] Example 2: Annotation of genomic regions of interest
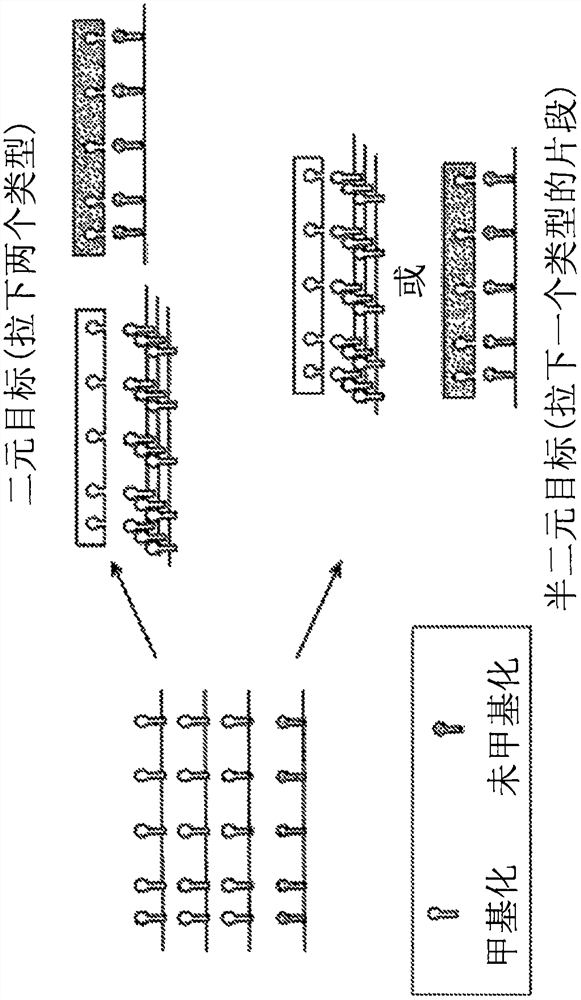
[0304] right through Figure 4 Genomic regions of interest identified by the process outlined in . Specifically, selected genomic regions of interest are aligned to a reference genome to determine a number of alignment positions. The alignment position information of each selected target genome region is collected, and the alignment position information includes: chromosome number, start nucleotide base, end nucleotide base and genome annotation of the given genome region. Target genomic regions are located in introns, exons, intergenic regions, 5'UTRs, 3'UTRs, or control regions such as promoters or enhancers. The number of genomic regions of interest within each genome annotation was counted and plotted in the graph provided in Figure 11. Figure 11 also compares the number of selected genomic regions of interest (black bars) or randomly selected genomic regions (gray bars) within each genome annotation.
[030...
example 3
[0306] Example 3: Cancer assay panel for detection of cancer and cancer type
[0307] Samples used for genomic region selection: DNA samples for this work were obtained from various sources.
[0308] The Circulating Cell-Free Genome Atlas study (CCGA; Clinical Trial.gov identifier (NCT02889978)) was a prospective, multicenter, case-control, observational study with longitudinal follow-up. Unidentified individuals were collected from approximately 15,000 participants at 142 sites. Identified biological samples. Samples were selected to ensure a pre-specified distribution of cancer types and non-cancer in each population, and cancer and non-cancer samples were frequency-age-matched by sex.
[0309] The Cancer Genome Atlas ("TCGA"; Clinical Trial.gov identifier NCT02889978) is a public resource developed by the National Cancer Institute (NCI) in collaboration with the National Human Genome Research Institute (NHGRI).
[0310] Isolated tumor cells (DTC) were obtained from Convers...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com