Small molecule inhibitor of ubiquitin-binding enzyme E2T
A technology of ubiquitin-conjugating enzymes and ubiquitination, which is applied in the field of small molecule inhibitors of ubiquitin-conjugating enzyme E2T, and can solve the unreported problems such as the association between UBE2T and RACK1
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
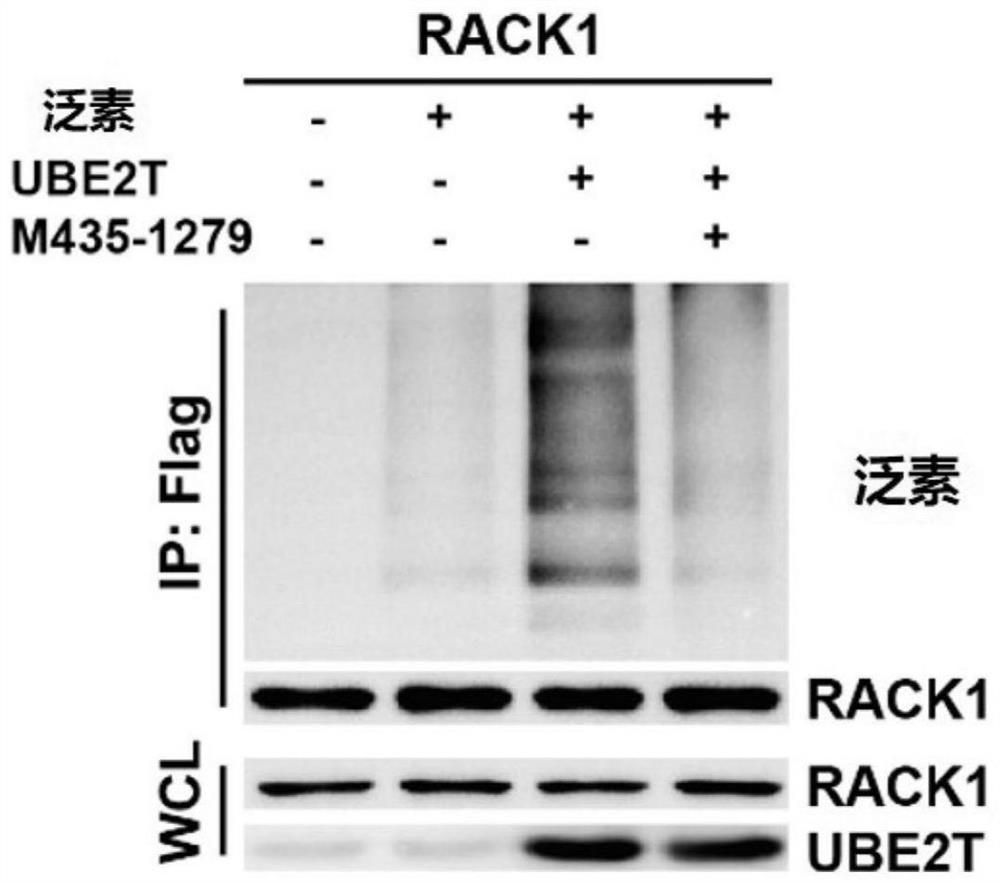
[0063] Example 1: M435-1279 binds and inhibits the activity of UBE2T
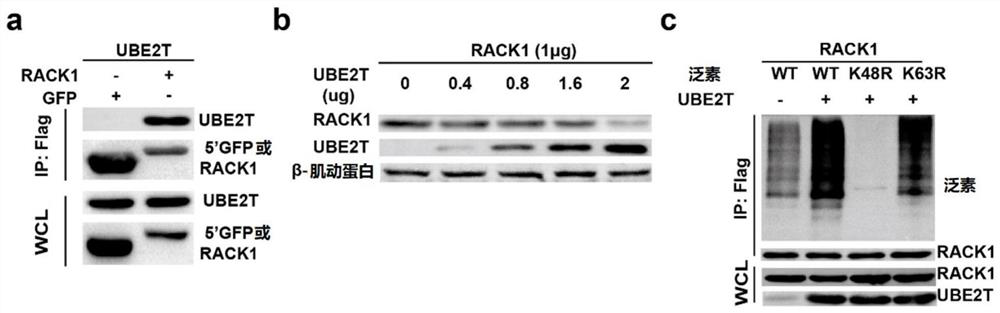
[0064] Flag-UBE2T and HA-RACK1 were transfected into HEK-293T cells, and 5'GFP (Flag-tagged GFP) was used as a control to verify the interaction between UBE2T and RACK1 by co-immunoprecipitation experiments ( figure 1 a), and Western blotting was performed by transfecting a fixed amount of RACK1 and different amounts of UBE2T. It was found that the more UBE2T was transferred, the lower the protein level of RACK1 was, proving that UBE2T can degrade RACK1 ( figure 1 b). Transfection of Flag-RACK1, untagged UBE2T, and HA-tagged different mutated ubiquitin chains into HEK-293T cells revealed that the K48R mutated ubiquitin chains could not be ubiquitinated, so the effect of UBE2T on RACK1 was dependent on (mediated degradation) of the K48 chain ( figure 1 c).
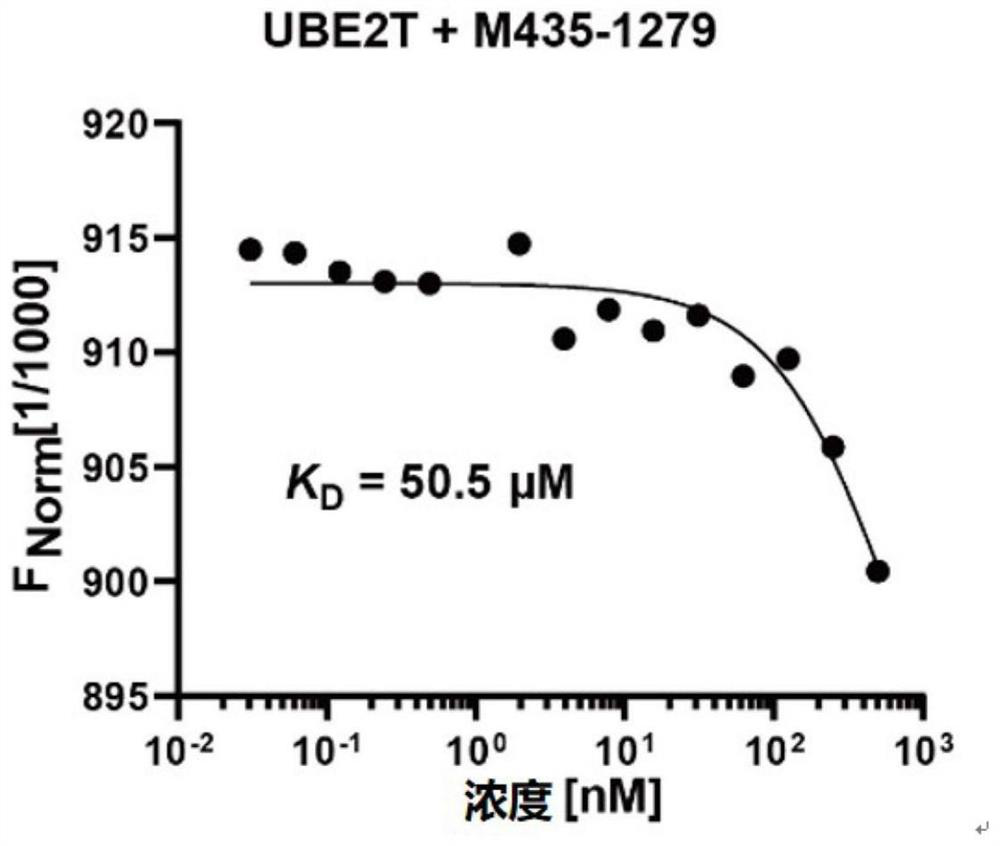
[0065] By microthermophoresis experiments ( figure 2 ) to test the binding ability of UBE2T and M435-1279, the KD value is 50.5 micromolar.
[006...
Embodiment 2
[0067] Example 2: M435-1279 in the treatment of gastric cancer
[0068] Clonogenic experiments ( Figure 4 a) Proved that the ability of colony formation was significantly weakened after adding M435-1279 to HGC27 and AGS in gastric cancer cells. Transweel experiment ( Figure 4 b) It proves that after adding M435-1279 to HGC27 and AGS in cancer cells, the invasion ability is significantly weakened. Gastric cancer cell MKN45 was used to plant subcutaneously in nude mice until the tumor grew to 80mm 3 When using M435-1279 for intratumoral injection (5mg / kg / day), and using DMSO as a control, it was found that the tumor growth rate in mice injected with M435-1279 was significantly slowed down ( Figure 4 c-e).
[0069] In conclusion, M435-1279 can significantly inhibit the progression of gastric cancer.
[0070]
[0071]
[0072]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com