Marker and model for predicting triple-negative breast cancer chemotherapy response
A triple-negative breast cancer, marker technology, applied in the field of precision medicine, can solve the problem of inability to guide TNBC, and achieve the effect of a good treatment plan
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0111] Example 1 Screening of TNBC gene diagnostic markers using mRNA expression data of genes
[0112] 1. Data set preparation
[0113] 1. Download the dataset GSE69031 (GPL571) from Gene Expression Omnibus (GEO), and then download the dataset TCGA-BRCA (GPL96, etc.) from TCGA. The data sets are gene chip data of breast cancer slices (Affymetrix platform GPL96, GPL571, etc.). A total of 171 cases were selected only from TNBC patients, of which TCGA-BRCA contributed 150 cases and GSE69031 contributed 21 cases.
[0114] 2. After removing the gene transcriptions with extremely low expression (the number of samples with non-zero expression does not exceed 10), miRNA and lncRNA are removed, and the common genes of the two data sets are selected, and the number of genes is 9524.
[0115] 3. Standardize the data of samples and genes step by step:
[0116] For each sample, the median of all gene expression levels was calculated separately, and the normalized expression of each sam...
Embodiment 2
[0135] Example 2 Cross Validation
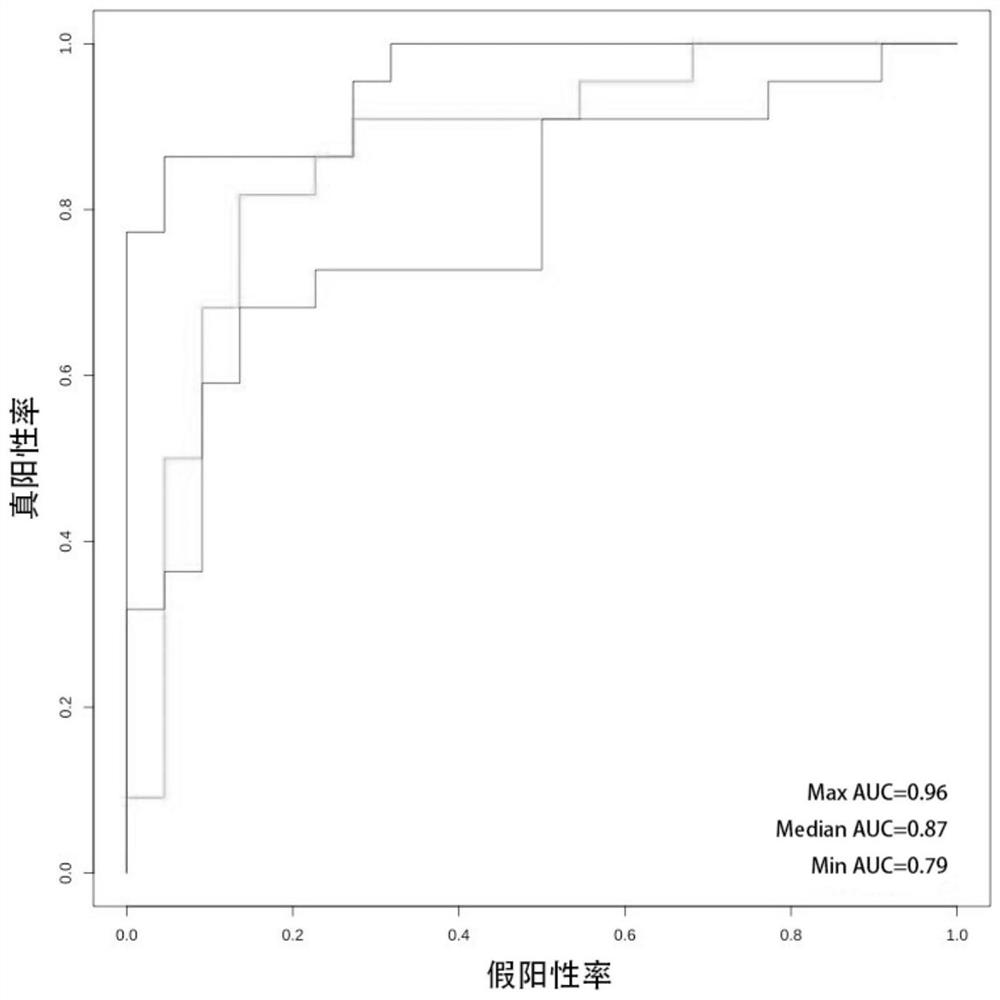
[0136] The data set in Example 1 is equally divided according to the population of the target variable, half of which is a training set and the other half is a verification set. The ROC curve analysis is performed on the model obtained in Example 1, and the AUC is calculated. This is repeated N (=20) times, and the statistical characteristics of AUC are calculated. see figure 1 As shown, the minimum value AUC=0.79, the maximum value AUC=0.96, and the median value AUC=0.87. Taking the median AUC value of the cross-validation as an indicator for evaluating the results of the model shows that the model provided by the present invention has excellence.
Embodiment 3
[0137] Example 3 Evaluation of any subset of markers
[0138] For the markers obtained in Example 1, take any subset (the subset includes K genes, K is a positive integer greater than or equal to 1 and less than 19), and evaluate the subset as a biomarker.
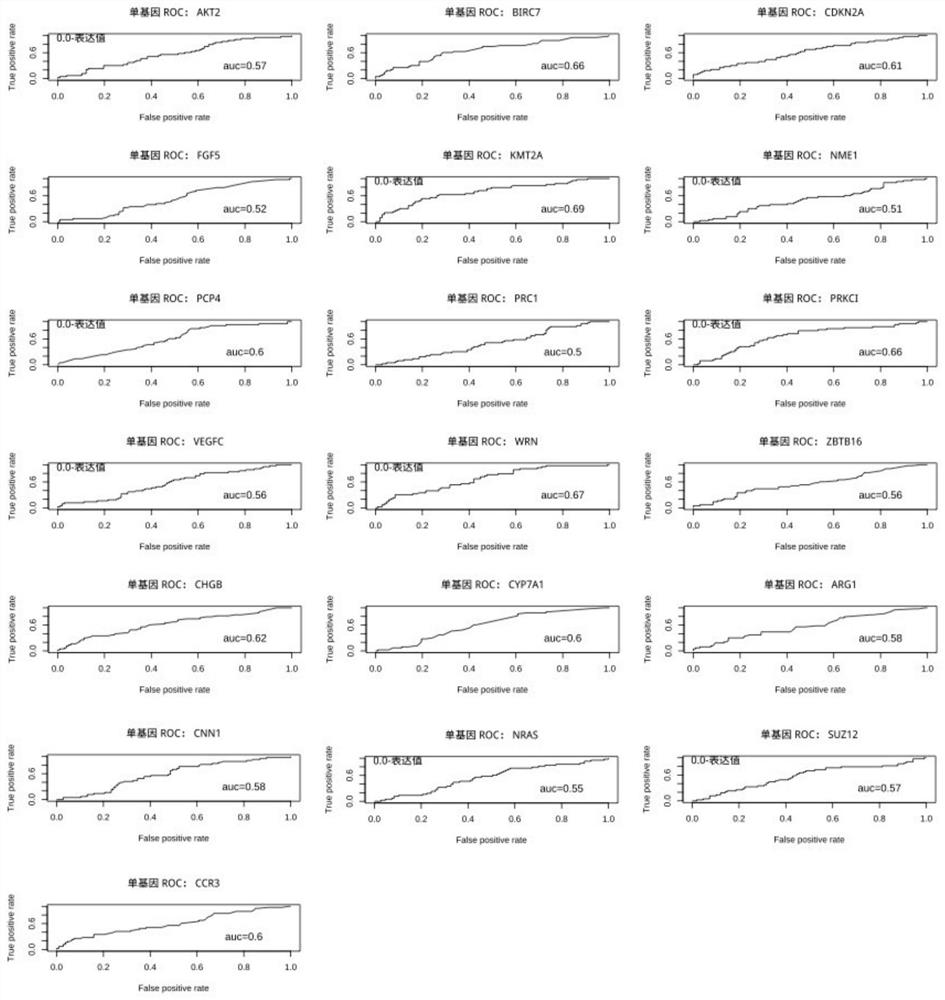
[0139] For the case of K=1, evaluate any single gene among the markers as a biomarker, draw its ROC curve and calculate the AUC, the results are shown in image 3 shown.
[0140] For the case of K ≥ 2, randomly select K genes among the markers, rebuild the model and perform cross-validation, draw the ROC curve and calculate the AUC, see some results Figure 4-7 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com