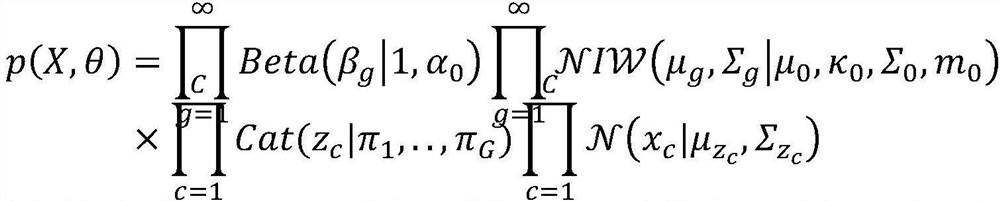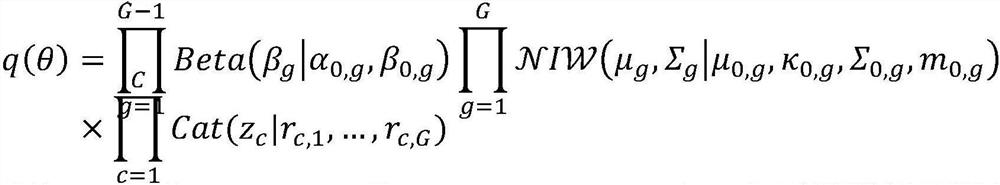Method for clustering metagenome sequences by using double-layer probability model
A probabilistic model and metagenomic technology, which is applied in the field of clustering metagenomic sequences using a double-layer probability model, can solve problems such as high pollution, low genome integrity, and inability of the probability model to fit features well, so as to improve clustering quality effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0031] The present invention will be further described in detail below in conjunction with specific embodiments, which are explanations of the present invention rather than limitations.
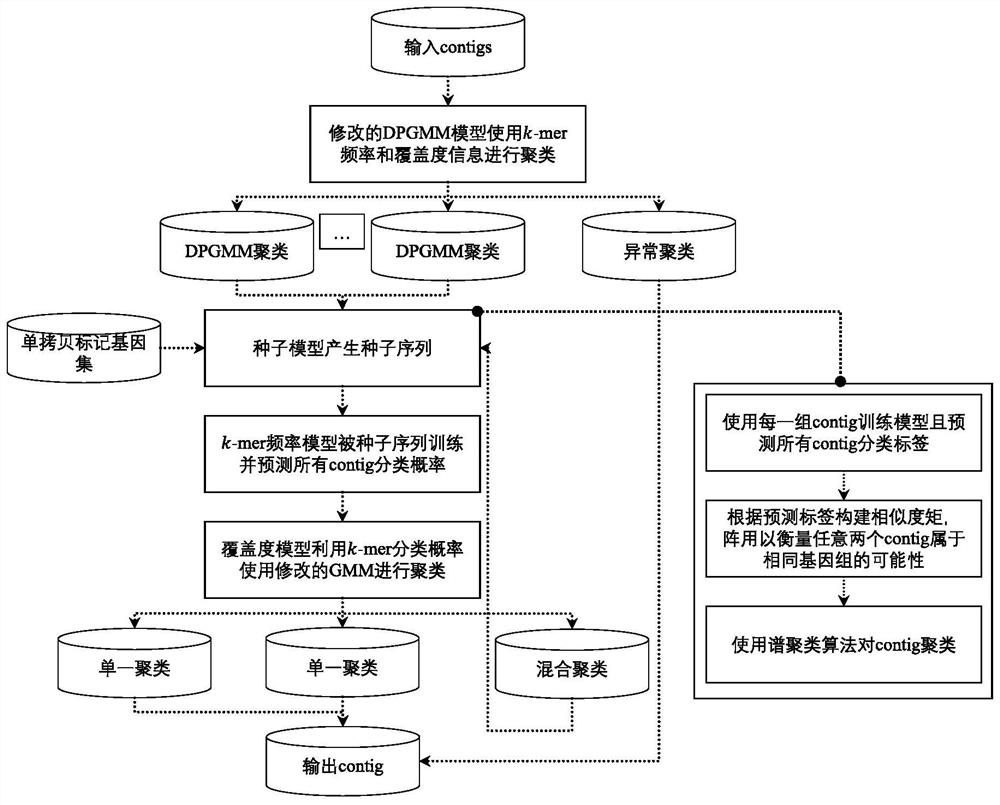
[0032] A method for clustering metagenomic sequences using a bilevel probability model, comprising the steps of:
[0033] P1, clustering all sequences in the initial metagenome using a first-level probabilistic model (DPGMM model) to obtain multiple primary clusters.
[0034] P11, obtain the k-mer frequency feature vector and coverage feature vector of all sequences from the initial metagenome, where k-mer represents an oligonucleotide with a length of k, and the value of k in the k-mer frequency feature vector is 4 ;
[0035] P12, merge the k-mer frequency feature vector and coverage feature vector into a single feature vector;
[0036] P13, use the Dirichlet process to construct the DPGMM model, and use the eigenvectors merged in P12 as input for clustering;
[0037] P14, use the variati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com