Coronavirus neutralizing effect protein and application thereof
A coronavirus and protein technology, applied in antiviral agents, virus/bacteriophage, peptide/protein components, etc., to achieve stable molecular structure and overcome poor specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
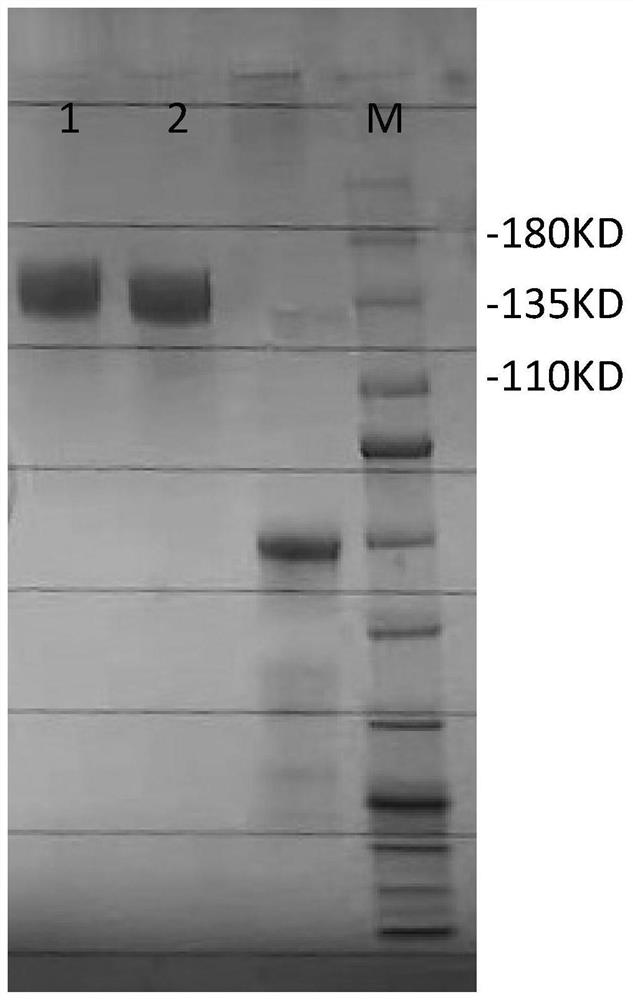
[0060] Embodiment 1, the preparation of ACE2 mutant protein C4-2
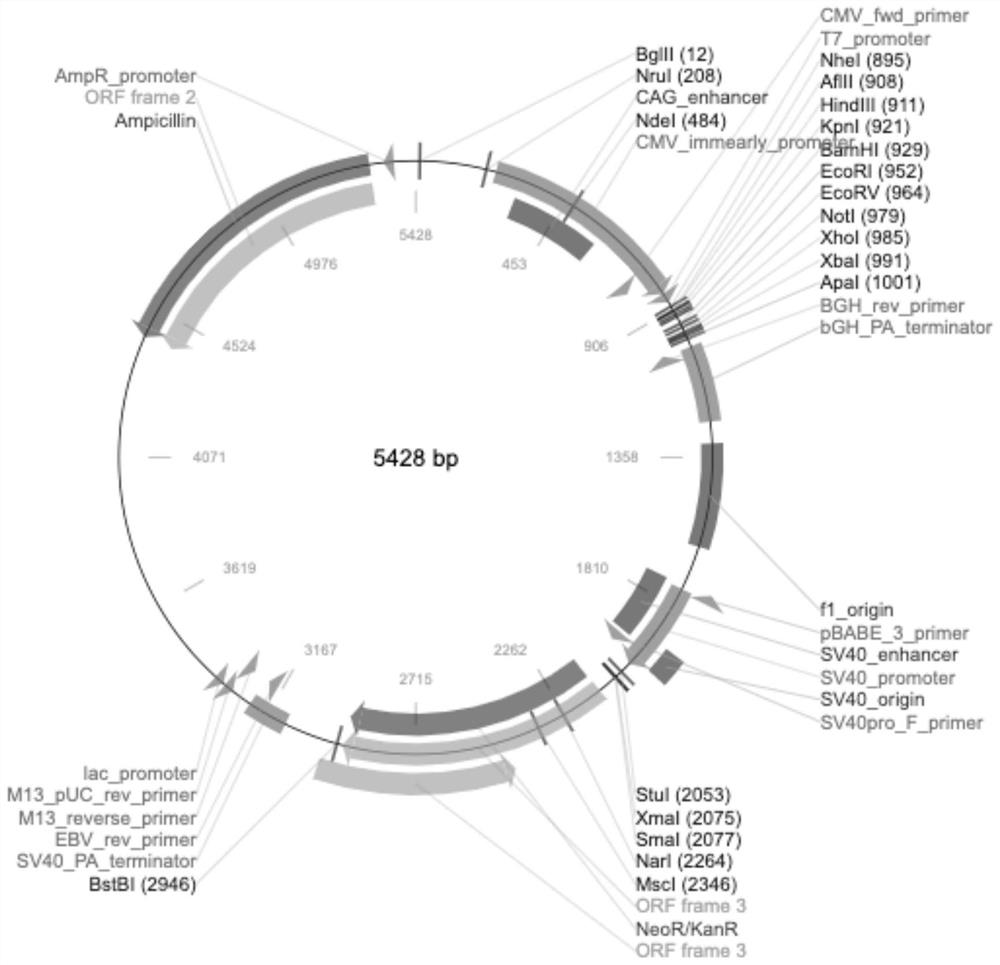
[0061] 1. Construction of recombinant expression vector pCDNA3.1 / C4-2
[0062] In the present invention, the 30th, 78th, 321st and 474th positions of the amino acid sequence of angiotensin-converting enzyme 2 (ACE2) are subjected to point mutations, and then ACE2 mutant protein C4-2 is obtained after fusion with the Fc segment of the mouse IgG antibody. The amino acid sequence is shown in SEQID No.1.
[0063] Design and artificially synthesize the eukaryotic expression DNA sequence (SEQ ID No.2) according to the amino acid sequence of the C4-2 protein, and add NheI (GCTAGC) and XbaI (TCTAGA) double restriction sites and homology arms were connected to the pCDNA3.1 vector with double restriction enzymes (NheI and XbaI) through Gibson Assembly, and then transferred into DH5α cells to amplify and extract the plasmid, and after sequencing, it was verified that it was correct The recombinant expression vector pCDN...
Embodiment 2
[0070] Example 2, ACE2 mutant protein C4-2 in vitro activity assay
[0071]1. Construction of cell lines expressing five S protein mutants of coronaviruses by lentiviral transfection
[0072] The five S proteins of SARS-CoV-2 come from: wild-type SARS-CoV-2, alpha, beta, gamma and delta epidemic strains of SARS-CoV-2. The amino acid sequence of the S protein from wild-type SARS-CoV-2 (ie Wuhan-Hu-1 strain) is the same as NCBI Reference Sequence: YP_009724390.1, and the corresponding coding gene sequence is NCBI Reference Sequence: NC_045512.2 (21563..25384) , NCBIGeneID is 43740568; the amino acid sequence of the S protein from the alpha epidemic strain has the following changes compared with YP_009724390.1: amino acid residues 69-70 are missing, amino acid residue 144 is missing, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H, the corresponding coding genes have the following changes compared with NC_045512.2 (21563..25384): 205-210 nucleotide deletion, 430-432 nucleotide ...
Embodiment 3
[0077] Example 3, Affinity Determination of ACE2 Mutant Protein C4-2 and SARS-CoV-2S Protein RBD
[0078] For the determination of typical affinity, the Biacore 2000 instrument was used in the experiment, and its detection principle is based on surface plasmon resonance (SPR), which can sensitively reflect the change of the refractive index on the chip surface. To study intermolecular interactions, one molecule is immobilized on the surface of the chip, and the other molecule flows across the surface in the form of a solution. The response value of SPR is proportional to the change of mass concentration near the chip.
[0079] In a typical test, the CM5 chip was chosen to measure the protein-protein interaction between soluble ACE2 protein and SARS-CoV-2 Spike-RBD. The CM5 chip is based on a covalent conjugation strategy with medium capacity and versatile properties. The running buffer was HBS-EP (Cytiva). RBD-his (Shenzhou: 40592-V08H105) (20ng / μl) was dissolved in sodium ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com