Protein family phylogenetic analysis method based on amino acid sequence alignment
A technology of sequence alignment and protein family, applied in the field of cluster analysis and biology, it can solve problems such as excessive calculation, achieve accurate results, improve clustering speed, and speed up.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
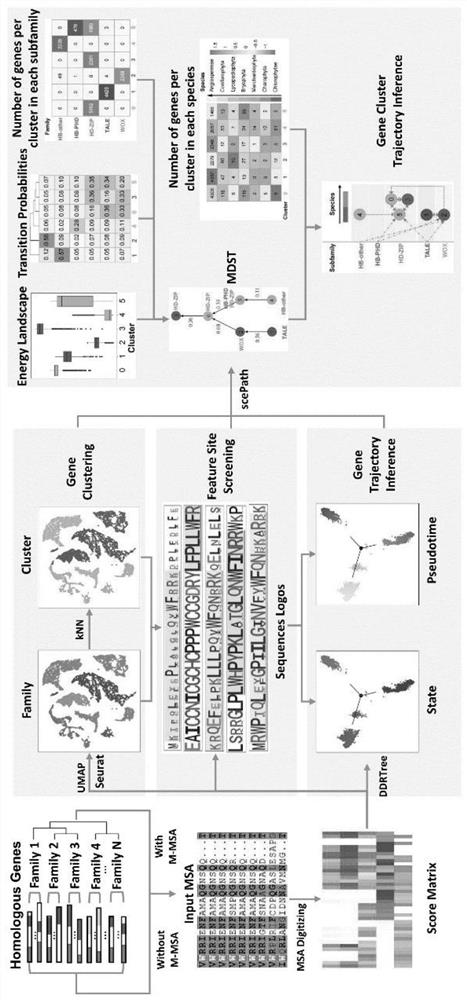
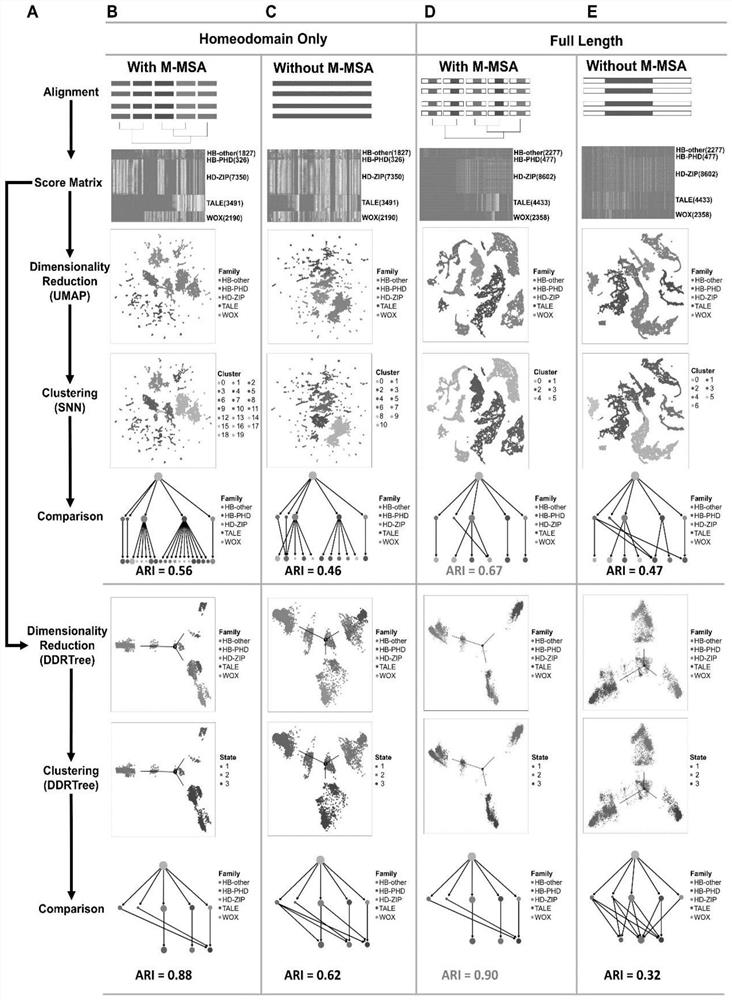
[0092] like Figure 1-2 As shown, the present embodiment provides a protein family phylogenetic analysis method based on amino acid sequence alignment, including:
[0093] 1. Amino acid sequence extraction of plant HB protein family: We retrieved the amino acid sequences of plant HB family full-length proteins (18147 in total) and Homeodomain (HD) (15184 in total) in the PlantTFDB database, and Its family annotation information.
[0094] 2. Obtain plant HB protein superfamily gene species annotation information: according to the degree of species development, according to the degree of species evolution, it is divided into angiosperms (Angiospermae), conifers (Coniferophyta), Lycopodiophyta (Lycopodiophyta) , Bryophyta, liverwort (Marchantiophyta), Charophyta and Chlorophytae. For dicots, the species information is divided into Asterids, Basal Magnoliophyta, Fabids, Malvids, Other Eudicots, Monocots (Monocots) and other plants (Other plants).
[0095] 3. Obtain the annotat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com