Oligonucleotide series for calculation of DNA chip
An oligonucleotide and computing chip technology, applied in the field of oligonucleotide sequences, can solve the problems of poor hybridization stability, unable to guarantee hybridization stability at the same time, no hybridization mismatch, weak signal, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
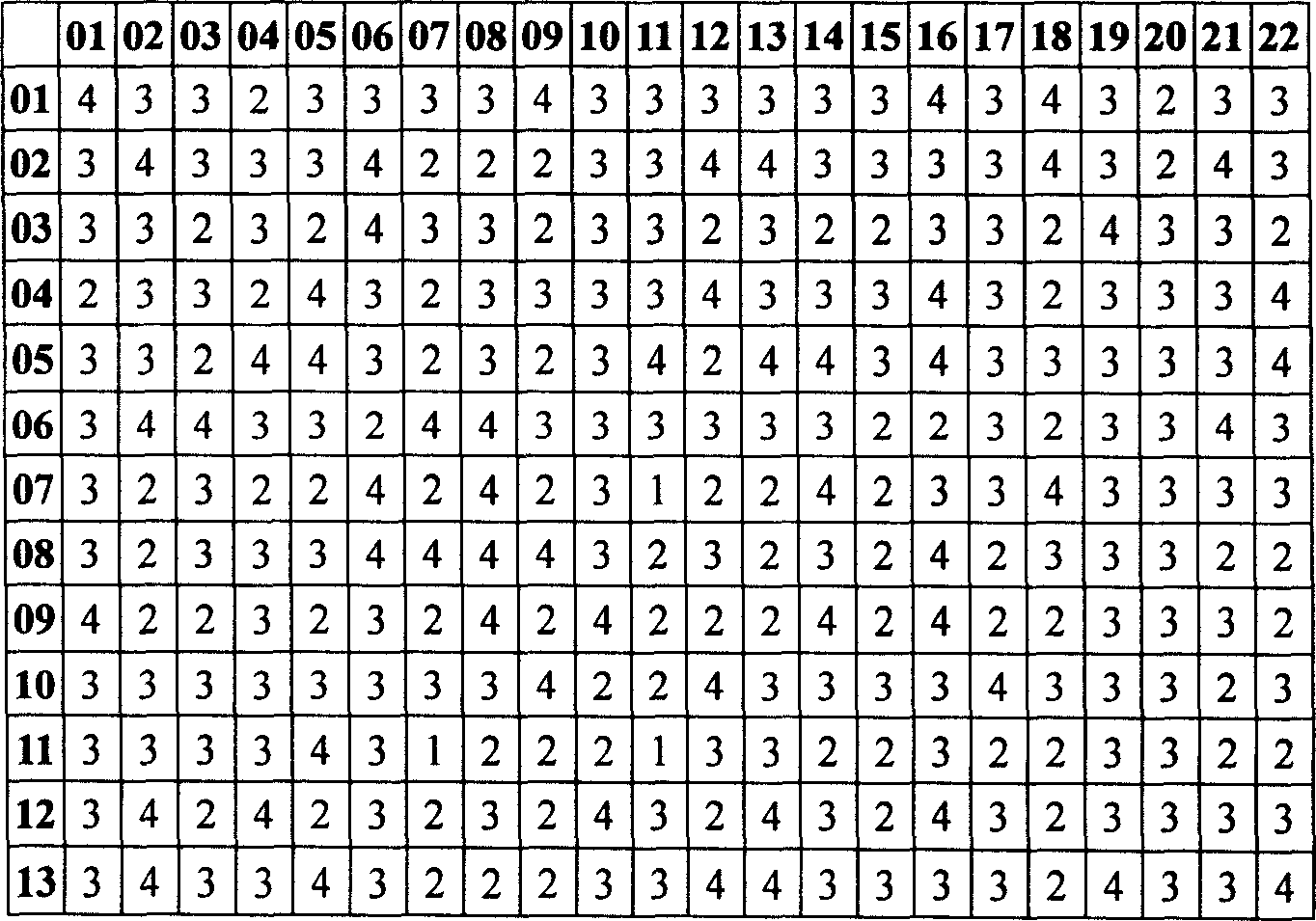
[0049]12-base variable parent sequence (n=22) pairing test results (comparison scores are expressed in matrix arrangement):
[0050] 01
[0051] 01
[0052] In the summary of the invention, the constraint condition (4) "the alignment score between the complete complementary sequences of any sequence A and any sequence B (A, B can be the same)" test results:
[0053]
[0054] 14
[0055] The above test results show that the selected 22 sequences all meet the constraint conditions (3) and (4) in the content of the invention in the description, that is, the comparison scores are not greater than 4.
Embodiment 2
[0057] A 4-variable SAT problem is encoded with the parent sequence, and arranged as a computing chip.
[0058] For a 4-variable SAT problem such as
[0059] F=(w∨x∨y)∧(w∨y∨z)∧(x∨y)∧(w∨y)
[0060] gcagtatccaca aggaactgagct
[0061] variable value
[0062] Z=0
[0063] The steps for the DNA computing chip to calculate this problem are as follows:
[0064] (1) Generate a DNA surface computing chip
[0065] ① The SAT question contains 4 variables (w, x, y, z) and 4 clauses, each variable takes a value of 1 or 0, then the complete data pool of the question contains 2 4 = 16 data, arranged in order of wxyz, 16 data are as follows: 0000, 0001, 0010, 0011, 0100, 0101, 011O, 0111, 1000, 1001, 1010, 1011, 1100, 1101, 1110, 1111
[0066] x
[0067] 0 0
[0068] ② Arrange the oligonucleotide sequences encoding the 8 states of the 4 variables of the above SAT problem on the chip according to the above array format to obtain an ...
Embodiment 3
[0074] The same 4-variable SAT problem as in Example 2 is encoded with the extended sequence, and arranged as a computing chip.
[0075] First pair: cagactagcctt aaggctagtctg
Second pair: ctctgattcgtc gacgaatcagag
Third pair: caaggacatacg cgtatgtccttg
Fourth pair: tcgctaaagggt accctttagcga
[0076] variable value
Sequence immobilized on chip (5'-3')
Fully paired probes (5'-3')
X=1
NH 2 -AAA AAA AAA AAA AAA cagactagcctt
FAM-aaggctagtctg
X=0
NH 2 -AAA AAA AAA AAA AAA aaggctagtctg
FAM-cagactagcctt
Y=1
NH 2 -AAA AAA AAA AAA AAA ctctgattcgtc
FAM-gacgaatcagag
Y=0
NH 2 -AAA AAA AAA AAA AAA gacgaatcagag
FAM-ctctgattcgtc
[0077] Z=1
NH 2 -AAA AAA AAA AAA AAA caaggacatacg
FAM-cgtatgtccttg
Z=0
NH 2 -AAA AAA AAA AAA AAA cgtatgtccttg
FAM-caaggacatacg
W=1
NH 2 -AAA AAA AAA AAA AAA tcgctaa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com