Detection and identification of human papillomavirus by PCR and type-specific reverse hybridization
a technology of reverse hybridization and detection method, which is applied in the field of detection and identification of human papillomavirus by reverse hybridization and type-specific reverse hybridization, can solve the problems of insufficient sensitivity and specificity of diagnosis by detection of hpv anti-bodies, and is not possible to diagnose hpv by culture, etc., and achieves rapid and reliable detection methods.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
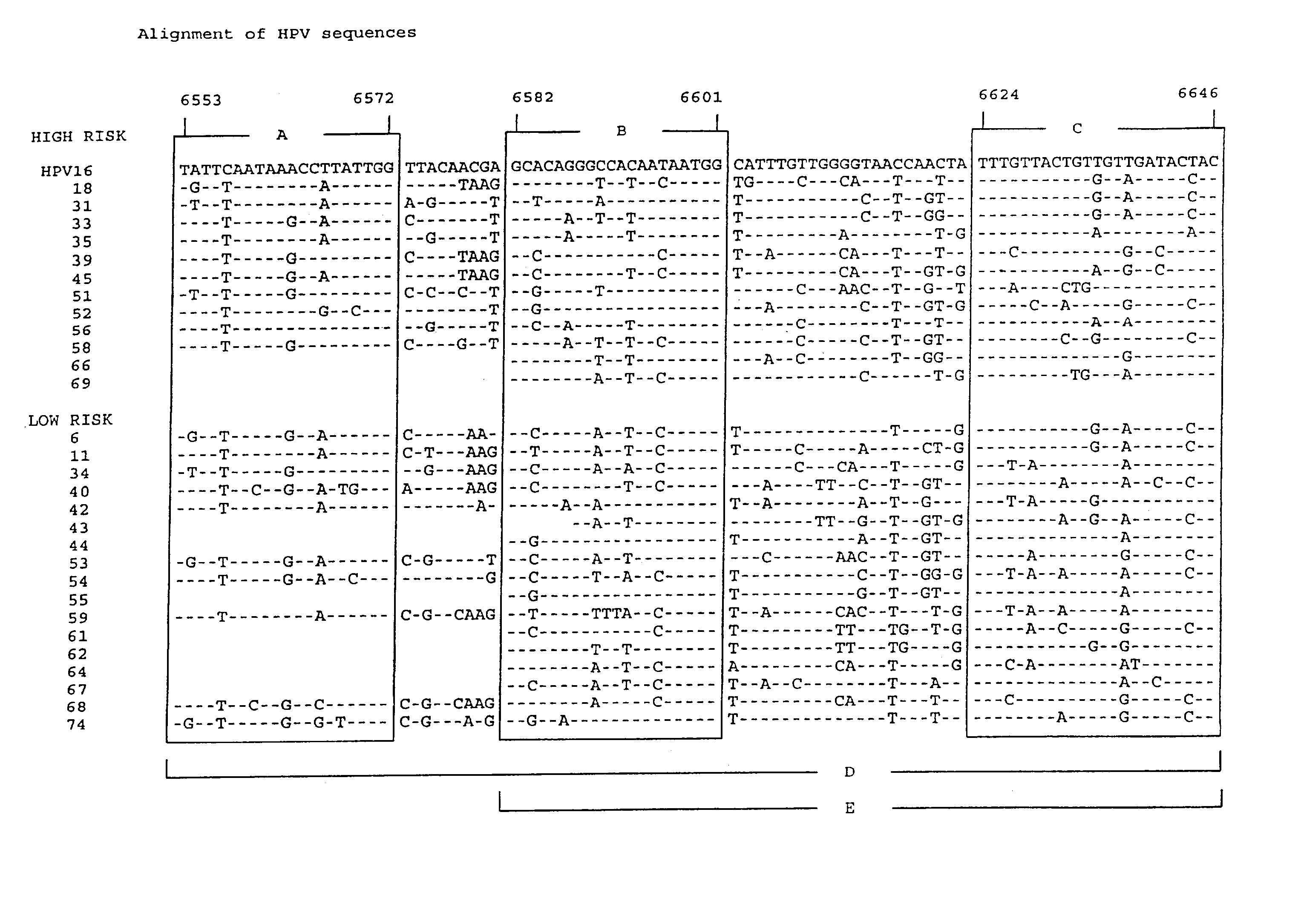
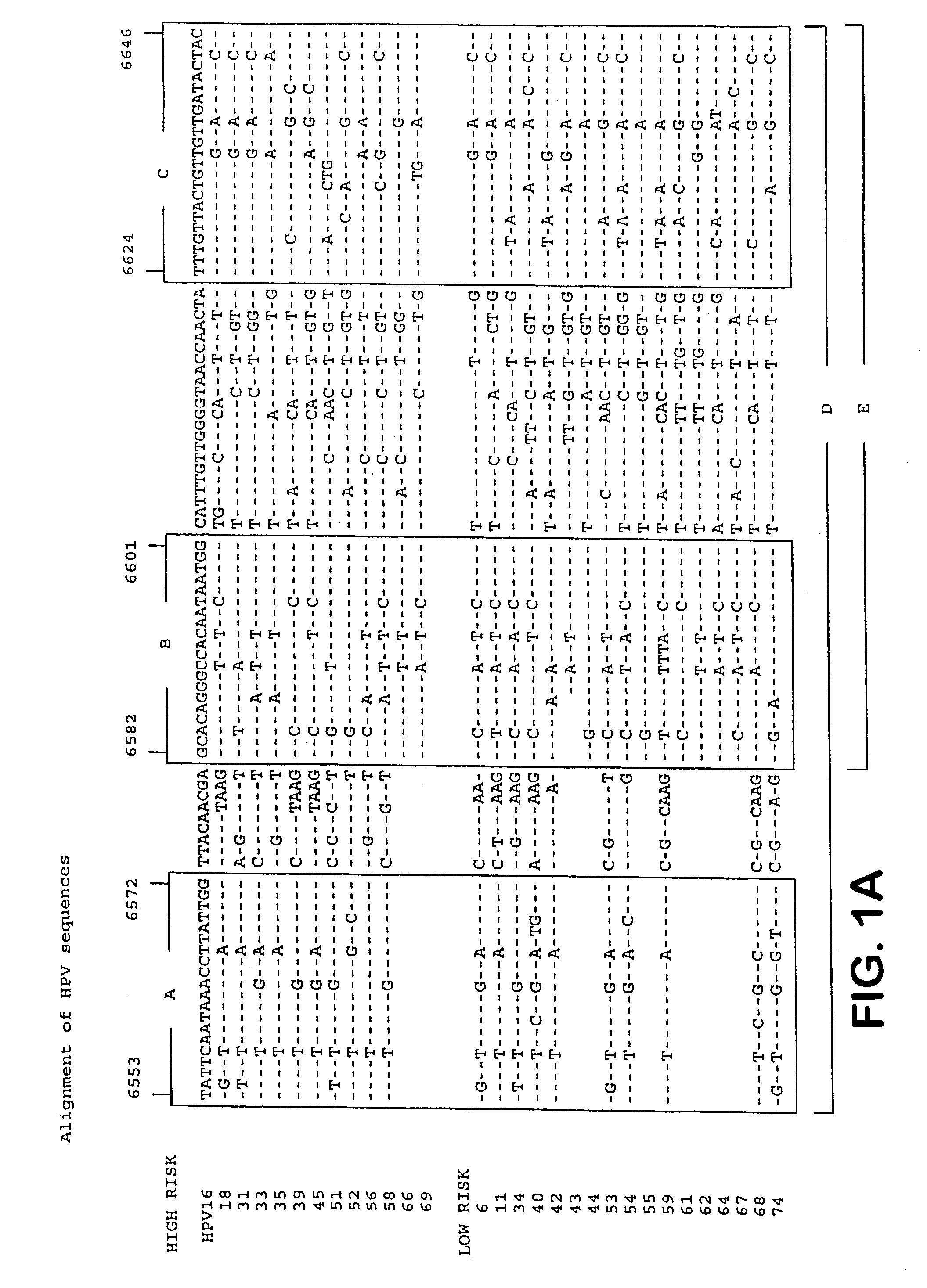
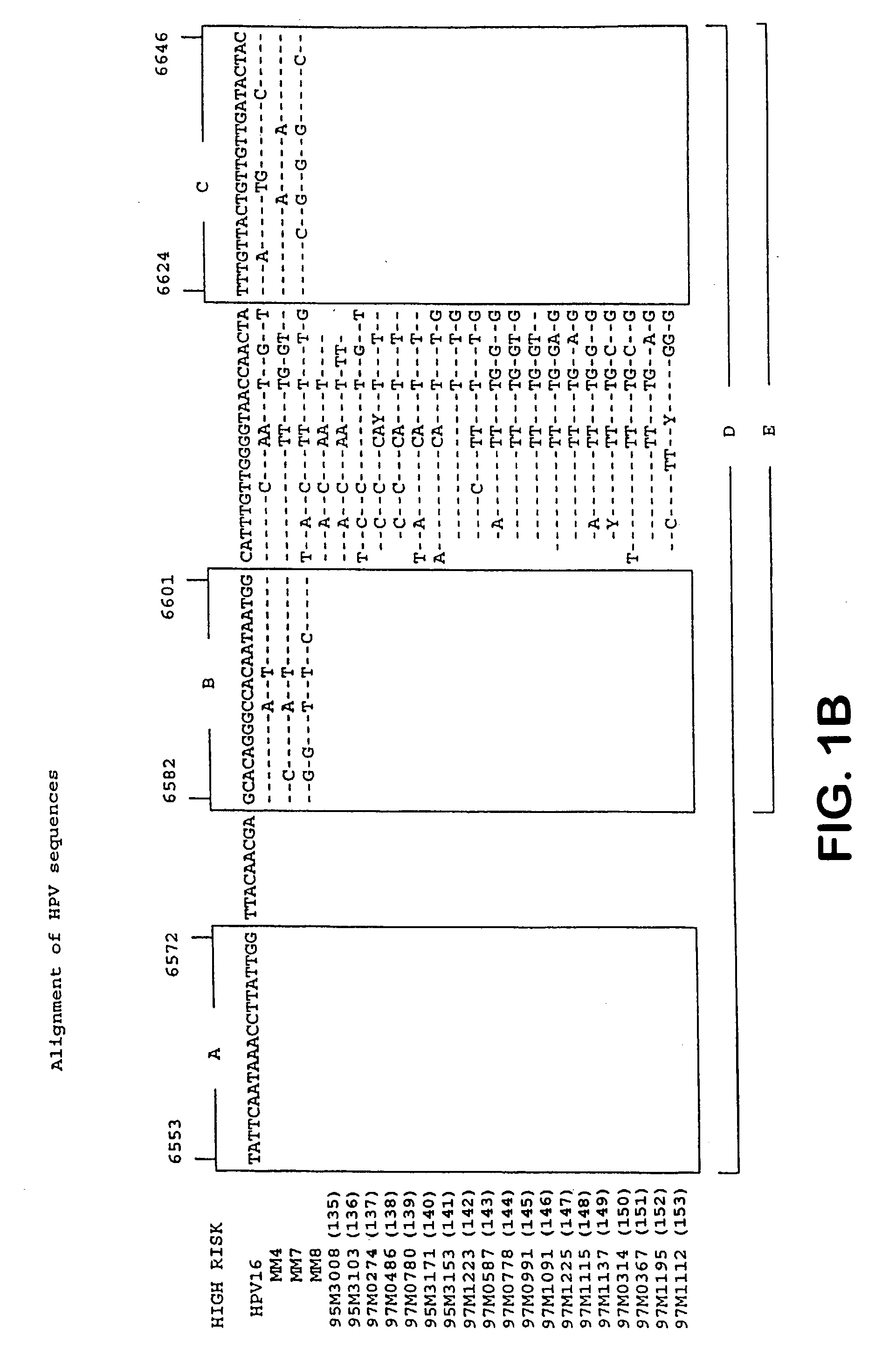
[0177] Example 1 describes the selection of semi-conserved regions in the L1 gene of the HPV genome, that permitted the development of a general PCR system. Degenerated primers were used for universal amplification of HPV sequences from different genotypes.
[0178] The present example describes the optimization of the primers aimed at these regions. Instead of degenerated primers, this study aimed at the development of several distinct and defined forward and reverse primers.
[0179] Materials and Methods
[0180] Alignments of L1 sequences were used to deduce PCR primers from the three regions A, B and C (FIG. 1). Primers were tested by PCR in different combinations on plasmids, containing partial or complete genomic inserts from the genital HPV types 6, 11, 16, 18, 31, 33, 35, 39, 43, 45, 51, 52, 53, 56, 57, 58, 59, 62, 64 and 67 as listed in table 2.
[0181] HPV DNA amplification was performed according to the following protocol. The final PCR volume of 100 .mu.l contained 10 .mu.l of HPV...
example 3
Identification of Different HPV Types by Analysis of a Small PCR Fragment Derived from the L1 region
[0187] Introduction
[0188] Identification of the different HPV genotypes may have great clinical and epidemiological importance. Current classification methods are for instance based on either type-specific PCR or sequence analysis of larger DNA fragments. Therefore, there is a clear need for a simple, rapid and reliable genotyping assay for the different HPV genotypes.
[0189] This assay should preferably be combined with the detection of HPV DNA, aiming at the same genomic region. Therefore, we aimed at the development of a screening assay to detect the presence of HPV DNA in clinical samples, and (in case of a positive screening result) the subsequent use of the same amplimer in a genotyping assay.
[0190] The theoretical requirements for such an assay would be as follows:
[0191] 1. The amplimer should be small, to allow highly sensitive detection and to permit amplification from formali...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Tm | aaaaa | aaaaa |
| melting temperature | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com