Enzyme-free isothermal exponential amplification of nucleic acids and nucleic acid analog signals
a nucleic acid analog and isothermal technology, applied in the field of detection of target nucleic acid molecules, can solve the problem of taking three hours to complete the amplification process, and achieve the effect of increasing the secondary structur
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of a Translator for Conversion of a Target Sequence to a Propagating Oligonucleotide
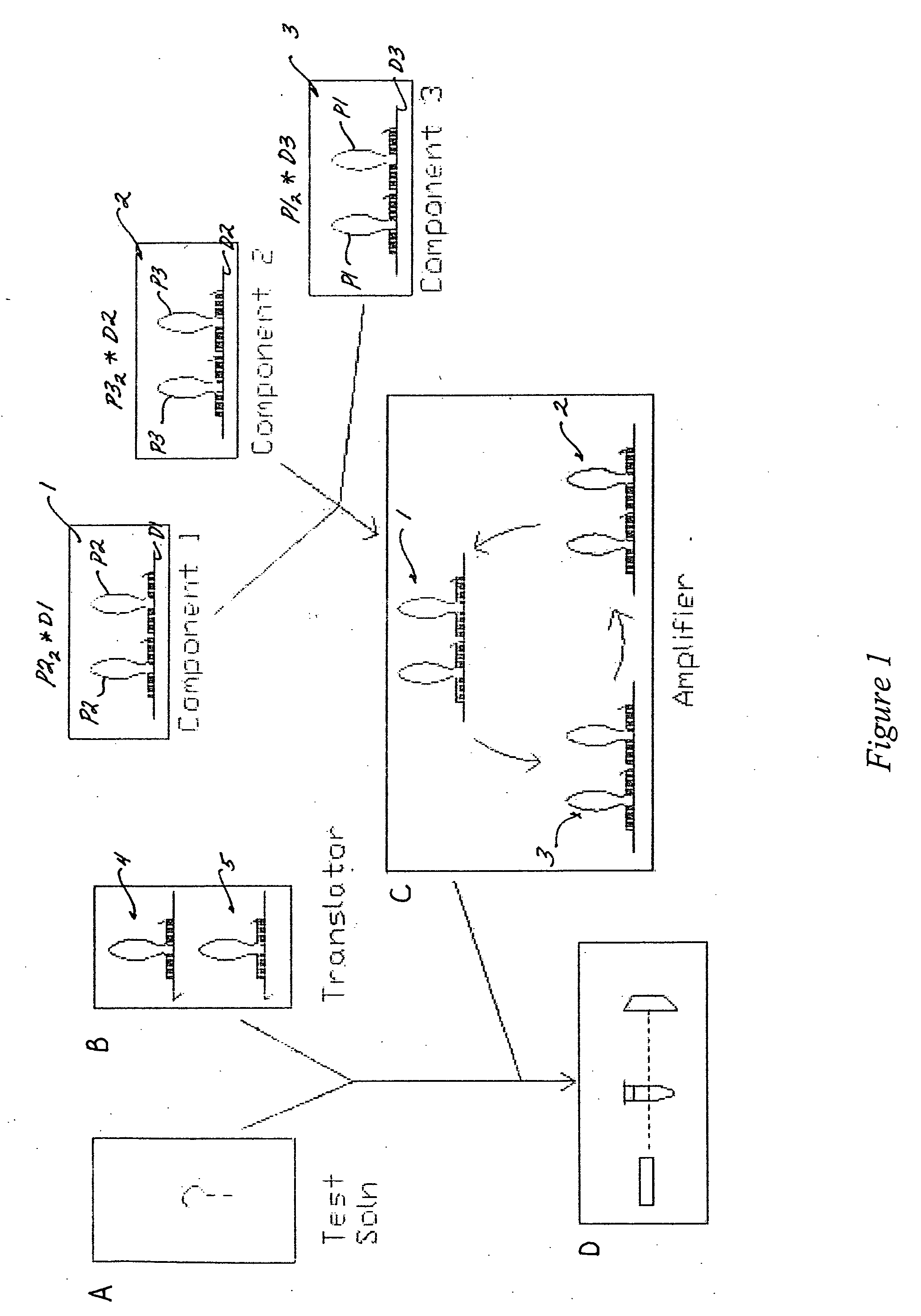
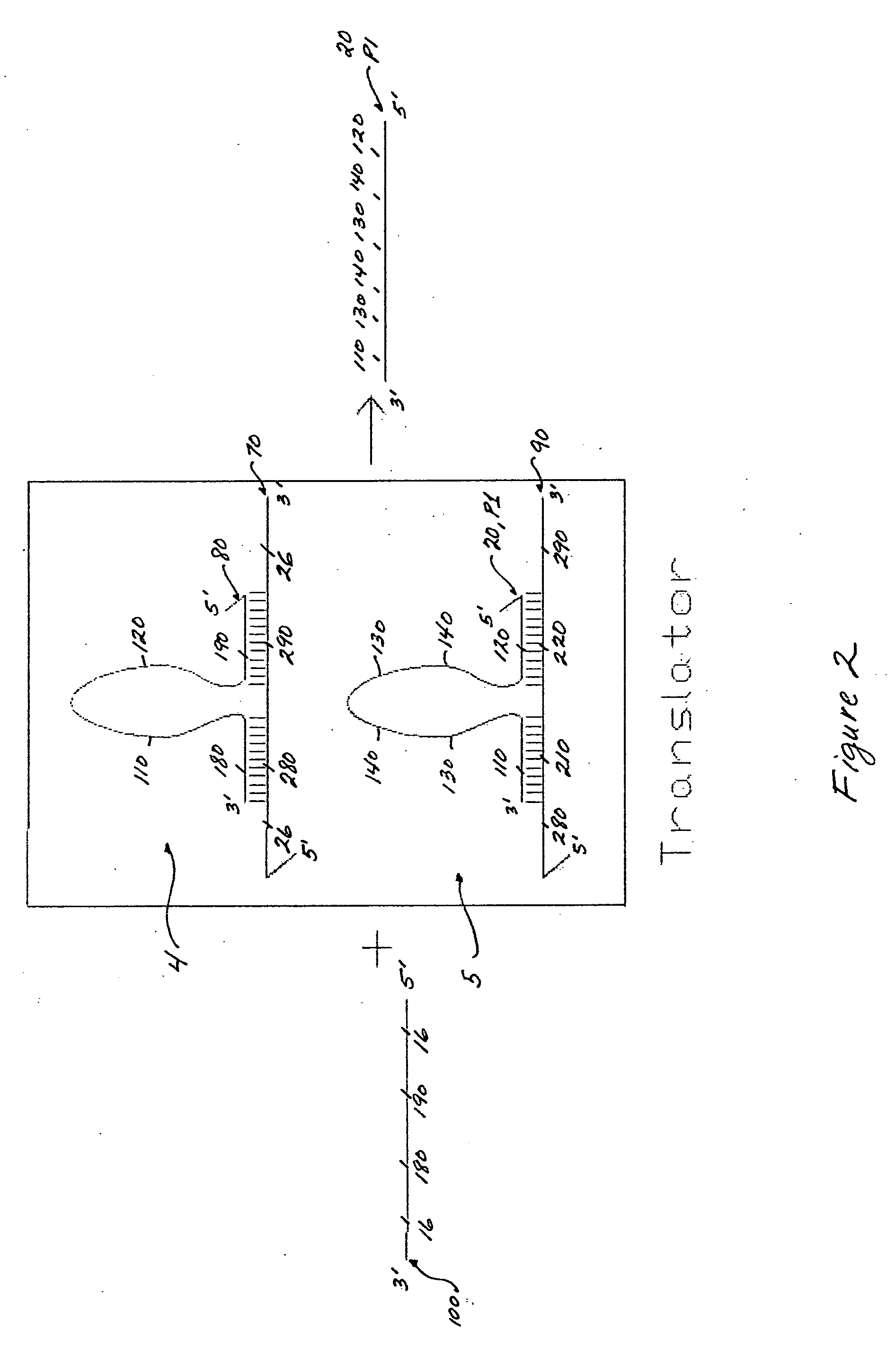
[0086] A translator is composed of two gain1 components (one propagating oligonucleotide to one damping oligonucleotide for each component; 1:1 ratio). The nucleotide and domain sequences of the translator gain1 components are fixed, relative to the desired target sequence and the gain2 components in the amplification system. Hence, the two gain1 components of the translator are modified as the target sequence changes (see FIG. 2). As such, there is a unique translator for every target sequence. The target sequence and its converted propagating oligonucleotide, which is dependent on at least one propagating oligonucleotide (P1, P2 or P3) of the amplification system, determine the sequence design of every oligonucleotide (70, 80, 90 and 20) used in the translator system (see FIG. 2).
[0087] The oligonucleotide that triggers the amplification system is one of the three propagating oligonuc...
example 2
Conversion of a Target Sequence to a Propagating Oligonucleotide in a Stoichiometric Ratio 1:1
[0089] The presence of the target sequence in solution is detectable only if the target sequence is first converted to a propagating oligonucleotide which is capable of initiating an amplification system or feedback loop mechanism (see FIG. 2). FIG. 2 shows a translator composed of two gain1 components 4 and 5. The first gain1 component 4 consists of a damping oligonucleotide 70 and a bound propagating oligonucleotide 80. The two oligonucleotides 70 and 80 are selectively hybridized together at sequence domains 180 and 190, in the 3′ to 5′ direction, of the propagating oligonucleotide 80, and 280 and 290, in the 5′ to 3′ direction, of the damping oligonucleotide 70. The damping oligonucleotide 70 contains at least one more binding domain 26, a toehold binding domain for the target sequence 8. The second gain1 component 5 also consists of a damping oligonucleotide 90 and a propagating oligo...
example 3
Binding of Propagating Oligonucleotide to a Damping Oligonucleotide Triggers Amplification
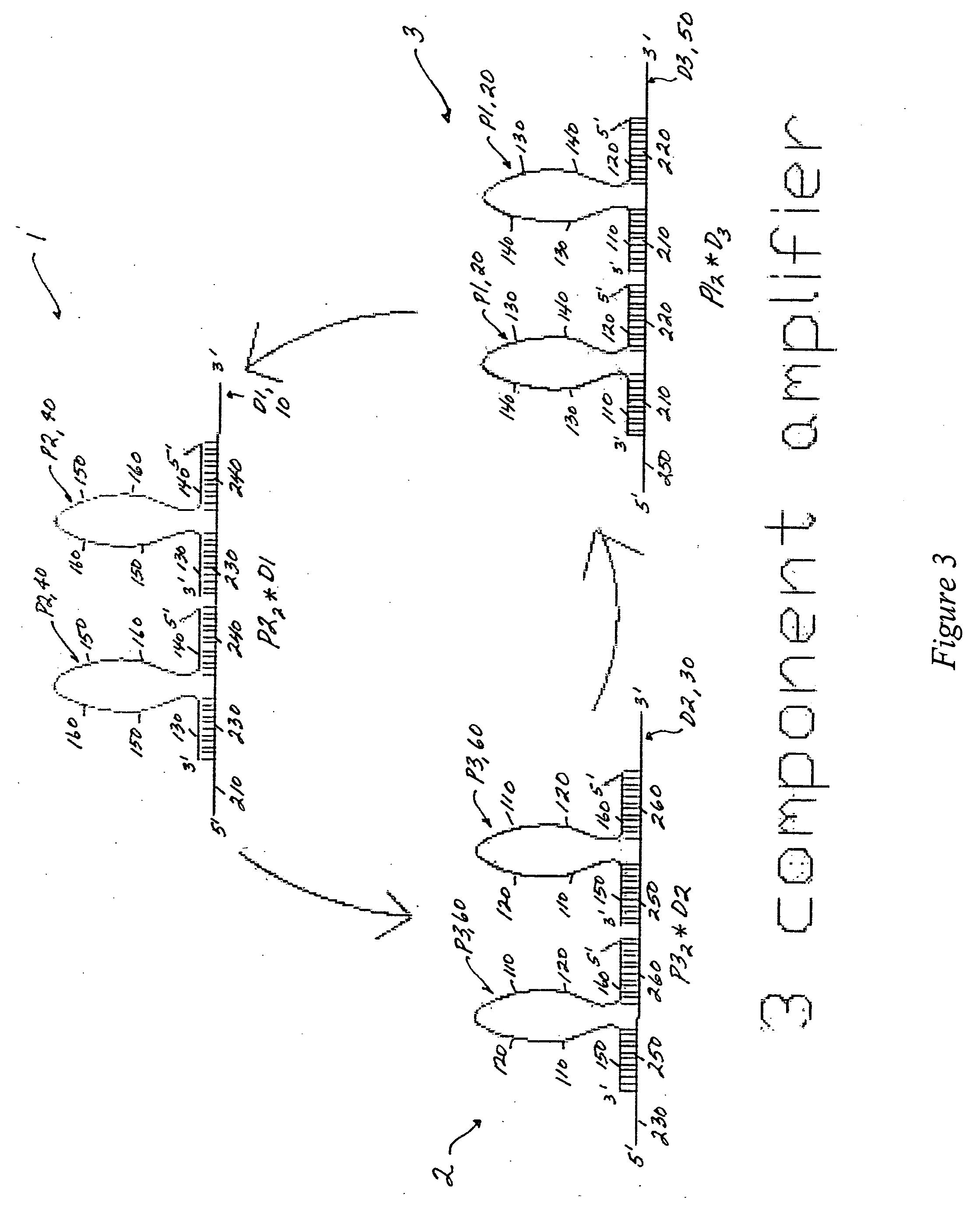
[0093] The converted target sequence (i.e., the initiator propagating oligonucleotide 20) can trigger an amplification system, as exemplified in FIG. 3 by a three gain2 component amplification system. The first cycle proceeds via the gain2 component 1, as illustrated in FIG. 5; the second, and third cycles function in substantially the same way. The translator output (initiator propagating oligonucleotide sequence 20; P1) is identical to the propagating oligonucleotides 20 (P1) of the gain2 amplifier 3, and is substantially complementary to the damping oligonucleotide 10 of the gain2 amplifier 1.
[0094] The three cycles of the amplification system can also be described in reactions sets (1), (2) and (3) below. The first cycle is represented by reaction (1); the second cycle by reaction (2); and the third cycle by reaction (3). The basic reaction set includes:
P1+P22*D1→P1*D1+2P2 (1)
P2+P32*D...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Time | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
| Resonance energy | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com