Method for identifying sub-sequences of interest in a sequence
a technology of sub-sequences and sequences, applied in the field of identification of sequences of interest, can solve the problems of difficult to identify meaningful or interesting sequences within a genome, the purpose of different parts of the genome is currently unknown, and the difficulty of traditional methods of identifying meaningful or interesting sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
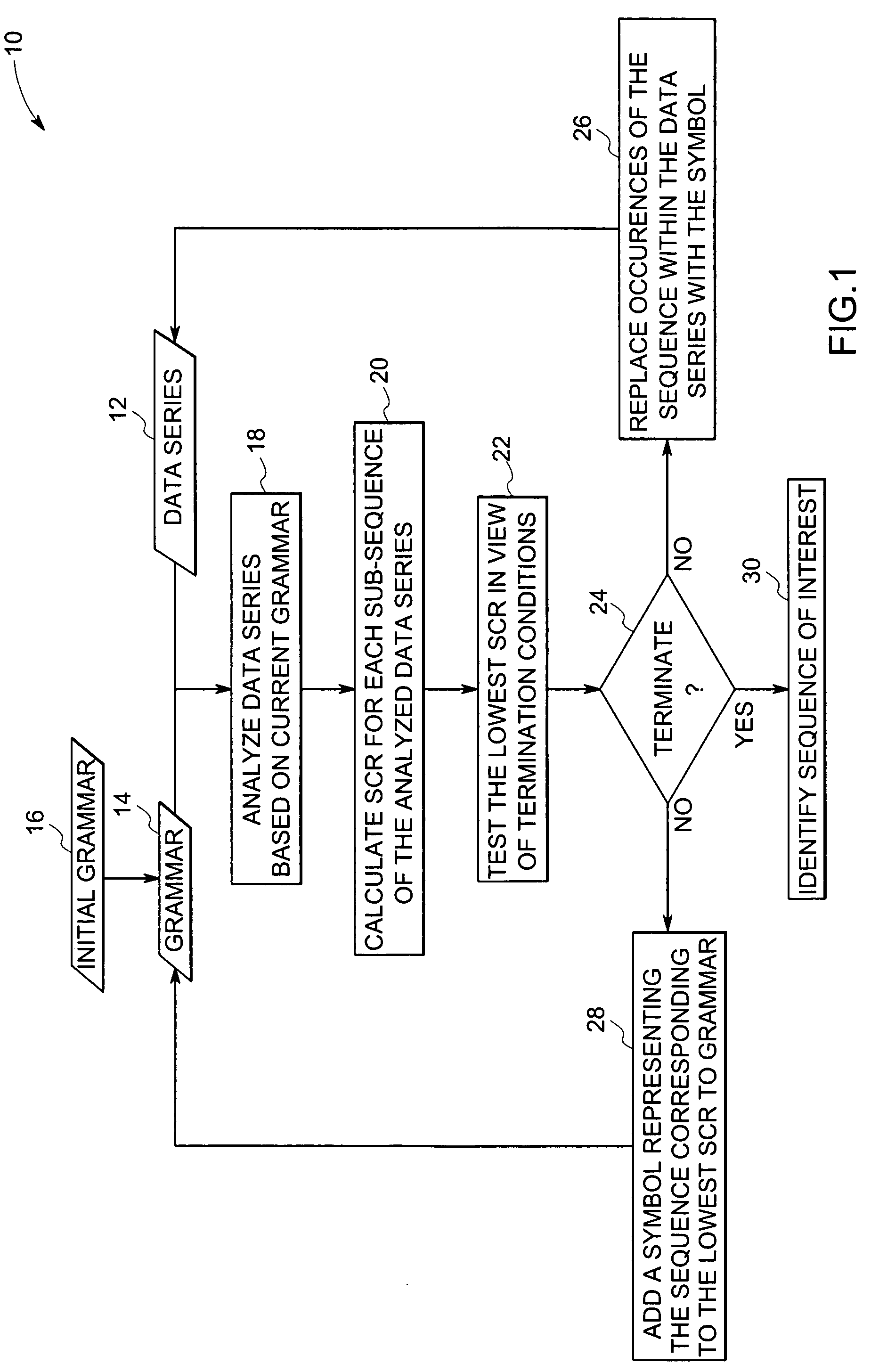
[0014] In many fields, such as genomic sequencing and analysis, it may be desirable to identify repetitive sequences, either to assist in compression and manipulation or to facilitate analysis. In particular, it may be desirable to identify such sequences in a computationally efficient manner. The techniques discussed herein address some or all of these issues.
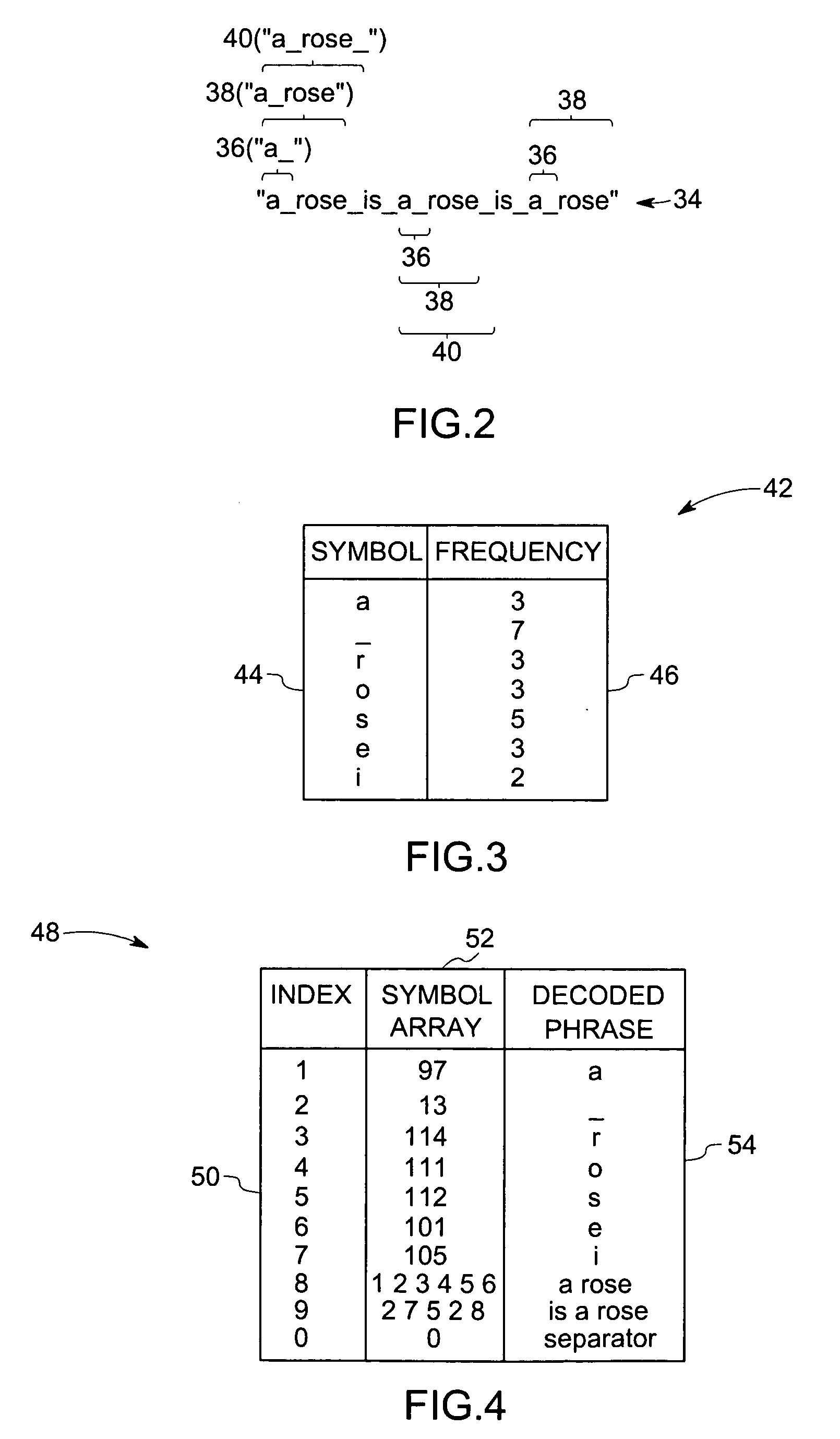
[0015] Turning now to the drawings, and referring to FIG. 1, a flow chart 10 depicts steps for identifying a sequence of interest, according to one aspect of the present technique. As suggested by the flow chart 10, a given data series 12 may be provided, within which may be one or more sequences of interest to be identified. The data series 12 may be constructed from a grammar 14. As will be appreciated by those of ordinary skill in the art, a grammar 14 may comprise terminals, i.e., uncombined symbols, and variables, i.e., combinations of terminals or terminal and other variables. For example, for a numeric data series 12, ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| symbol compression ratio | aaaaa | aaaaa |
| compression ratio | aaaaa | aaaaa |
| description length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com