Primer and probe design for efficient amplification and detection of HCV 3' non-translating region
a probe design and probe technology, applied in the field of oligonucleotides, can solve the problems of difficult study of rna, difficult identification of this agent, bartenschlager and lohmann
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
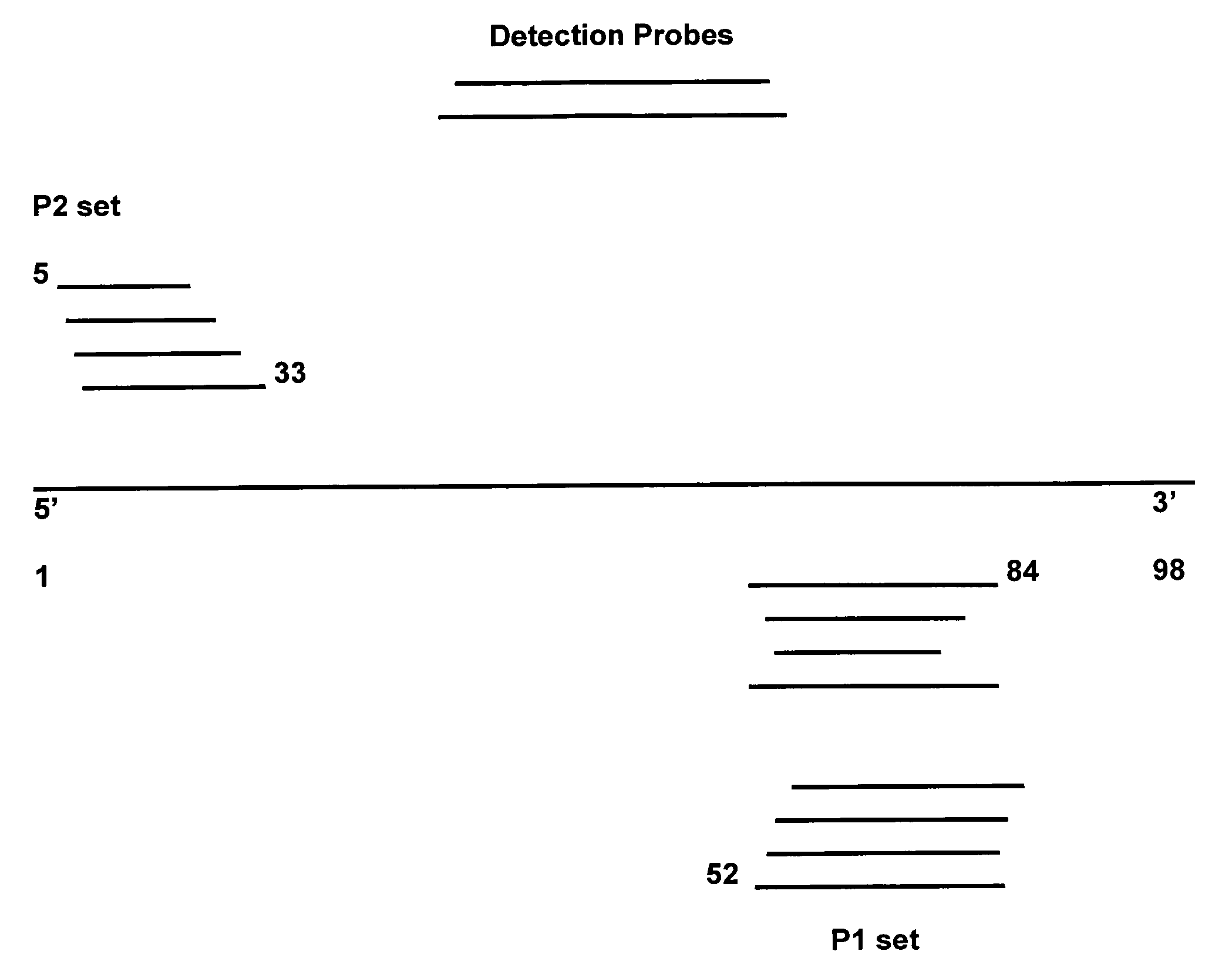
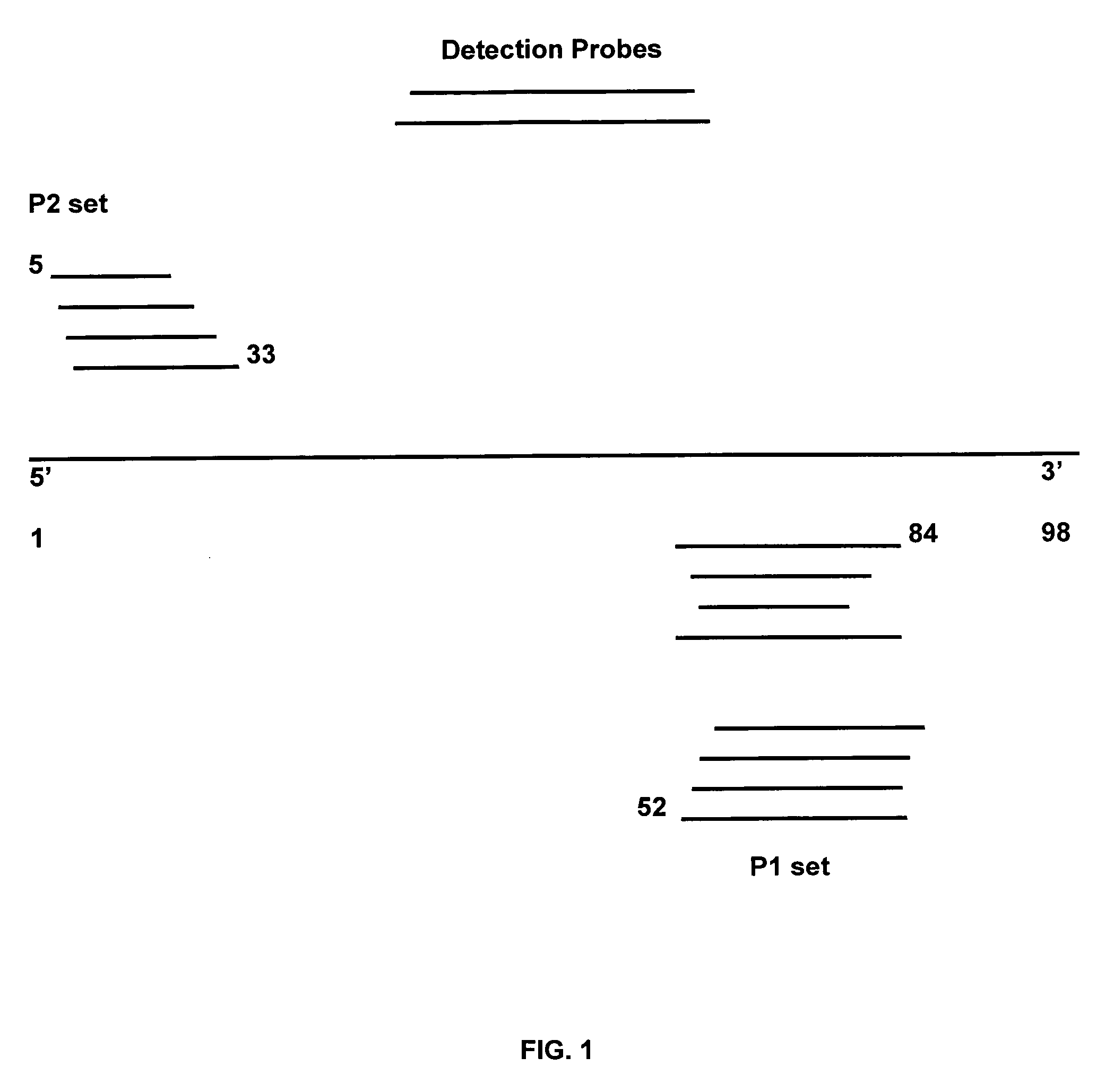
[0041] Within this application and particularly within the Examples, the primers utilized may be referred to as P1 primers or P2 primers. This terminology simply indicates that, within a NASBA or TMA reaction in these examples, the P1 primer is the primer having a T7 promoter sequence attached to its 5′ end (promoter primer); the P2 primer does not. It is not limiting terminology.
TMA Amplification and Detection of 3′ HCV
[0042] To evaluate the usefulness of various primers in HCV amplification, standard VIDAS Probe D2 qHCV assay conditions were used as follows. For lysis, 0.5 ml sample (such as EDTA-plasma, or in vitro RNA transcript diluted in base matrix) was mixed with 0.4 ml urea-based lysis buffer, incubated 67.5° C. (20 min) and cooled to room temp (20 min). During these incubations the released nucleic acid was captured by capB probe (SEQ ID NO: 19), which was linked to magnetic beads by a polyA tail. Complexes were washed and resuspended in RAR buffer (see Table 1). RNA ta...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Nucleic acid sequence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com