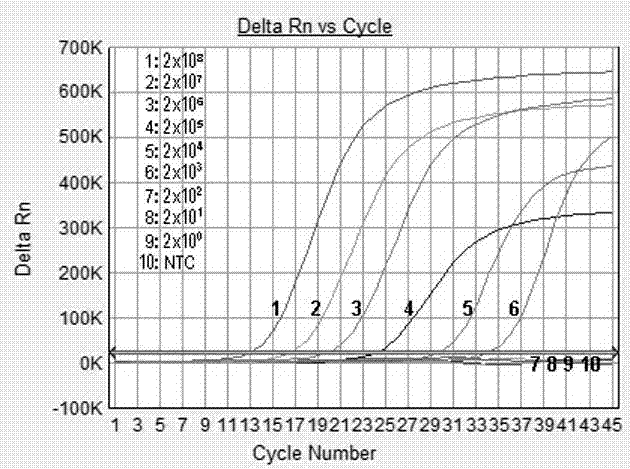
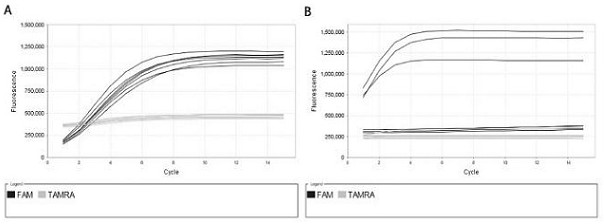
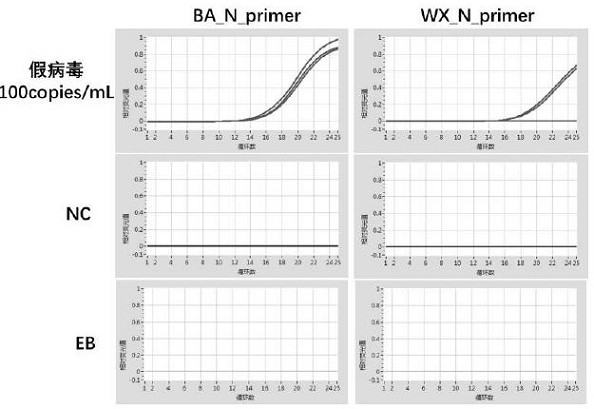
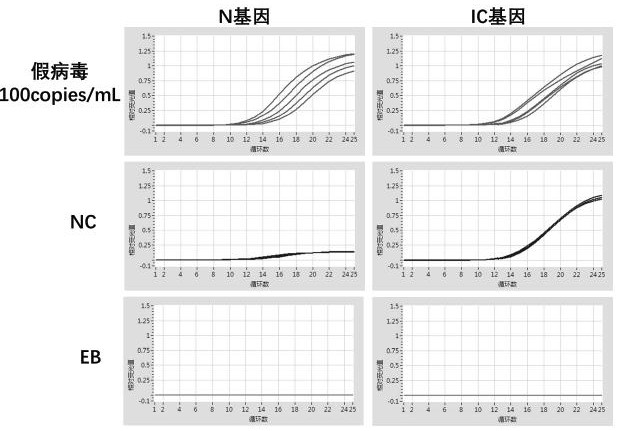
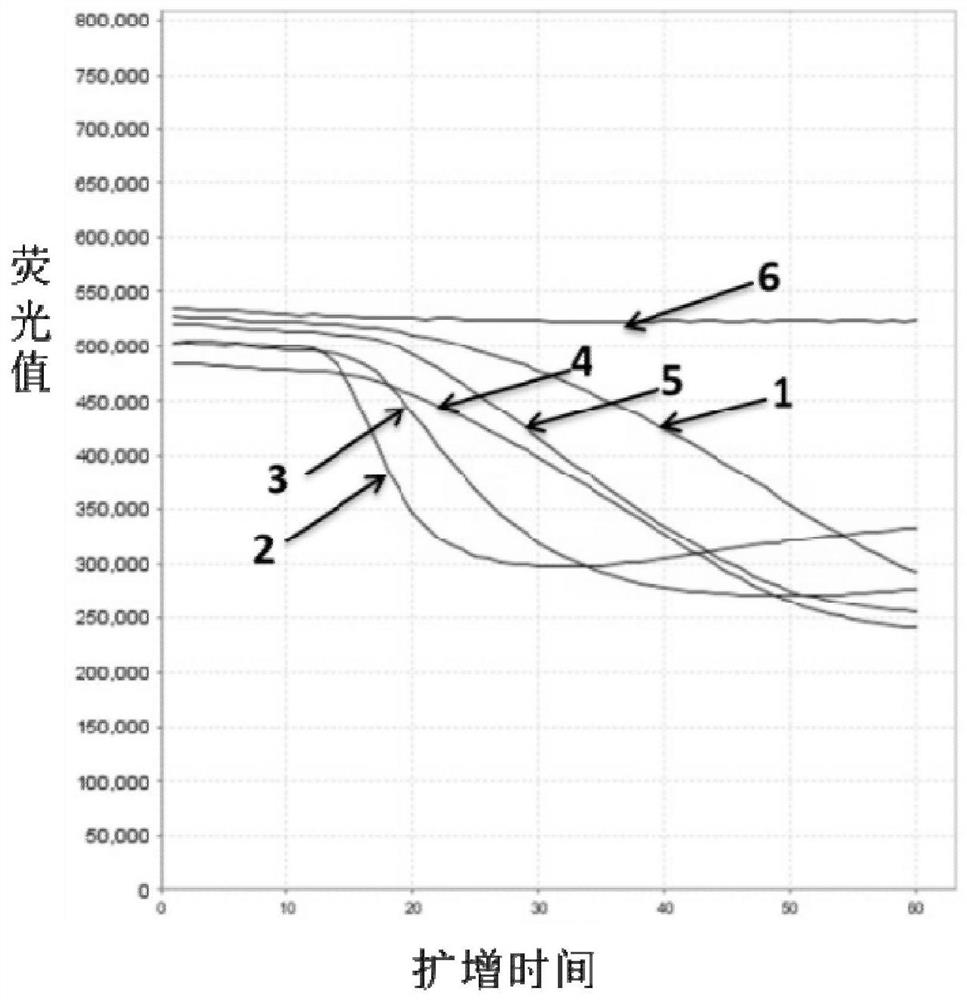
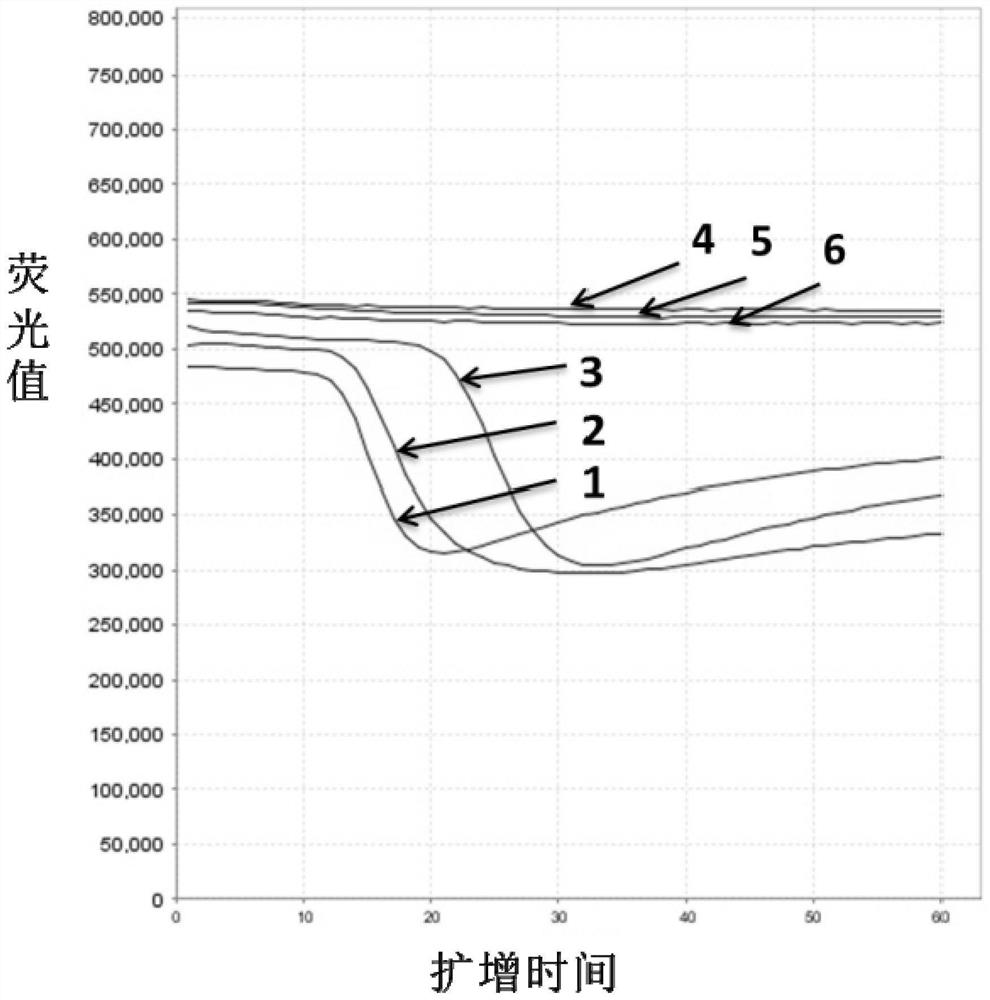
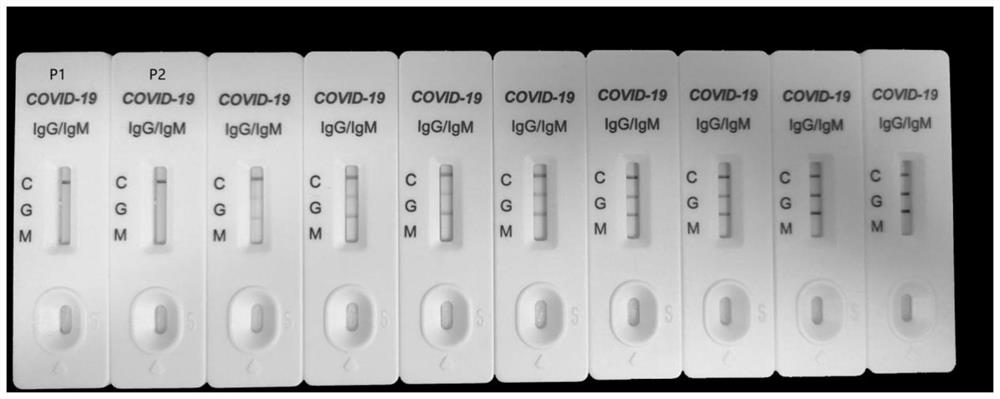
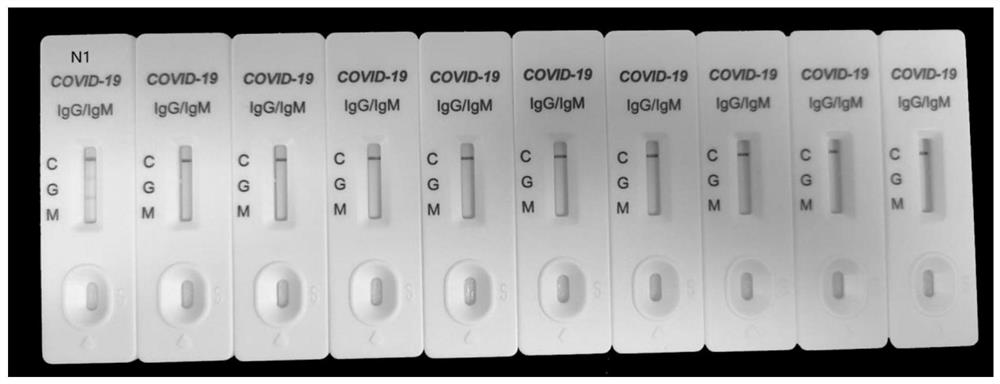
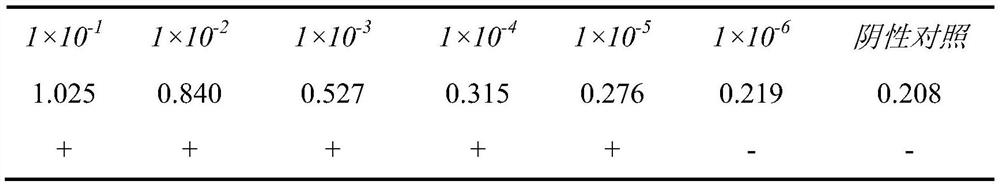Patents
Literature
37 results about "NASBA" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Nucleic acid sequence based amplification (NASBA) is a method in molecular biology which is used to amplify RNA sequences.
Norovirus real-time isothermal amplification detection kit, its primers and probe
InactiveCN102965452AAvoid inconvenienceStrong specificityMicrobiological testing/measurementMicroorganism based processesDiseaseMicrobiology
The invention relates to an enteropathogen rapid detection technology based on real-time nucleic acid sequence-based amplification (NASBA). Specifically, the invention provides a Norovirus real-time isothermal amplification detection kit, and a pair of primers and a molecular beacon probe thereof. The kit includes: a 2*real-time NASBA reaction solution containing the primers and the probe, a 5*enzyme mixed solution and a positive control template, a negative control and a blank control. The sequences of the primers and the probe are the sequences numbered as SEQIDNO:1-3, and the primers and the probe can specifically amplify and detect a Norovirus ORFs1 gene fragment. The kit provided in the invention has the characteristics of fastness, high efficiency, sensitivity and specificity, and real-time detection analysis, etc., and can be used in the fields of conventional detection and disease control and prevention in clinical practice and ports.
Owner:珠海国际旅行卫生保健中心
Group A rotavirus real-time isothermal amplification detection kit, primers and probe thereof
ActiveCN102965451AAvoid inconvenienceStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationDiseaseNucleic acid sequencing
The invention relates to an enteropathogen rapid detection technology based on real-time nucleic acid sequence-based amplification (NASBA). Specifically, the invention provides a group A rotavirus real-time isothermal amplification detection kit, a pair of primers and a molecular beacon probe. The kit includes: a 2*real-time NASBA reaction solution containing the primers and the probe, a 5*enzyme mixed solution and a positive control template, a negative control and a blank control. The sequences of the primers and the probe are the sequences numbered as SEQIDNO:1-3, and the primers and the probe can specifically amplify and detect group A rotavirus VP6 gene. The kit provided in the invention has the characteristics of fastness, high efficiency, sensitivity and specificity, and real-time detection analysis, etc., and can be used in the fields of conventional detection and disease control and prevention in clinical practice and ports.
Owner:SHENZHEN TOTAL TEST TECH
NASBA kit for detecting monkey pox virus and application thereof
InactiveCN103468830AQuick screeningGuaranteed specificityMicrobiological testing/measurementFluorescence/phosphorescenceAgricultural scienceQuarantine
The invention discloses an NASBA kit for detecting a monkey pox virus and the application thereof. The NASBA kit contains a composition used for detecting the monkey pox virus, the composition is composed of a primer pair and a probe, the primer pair is composed of two single-stranded DNA molecules shown in the first sequence and the second sequence in a sequence list, and the sequence of the probe is the third sequence in the sequence list. The NASBA kit is used for detecting the monkey pox virus, and the advantages of being simple, convenient, efficient, quick and peculiar are achieved. The NASBA kit can quickly screen the monkey pox virus and has great significance to screening port input diseases, technical support and strategic reserve are well prepared for preventing introduction of the diseases, and the demand of current health quarantine is met.
Owner:浙江国际旅行卫生保健中心
Kit and detection method for quickly detecting porcine transmissible gastroenteritis virus by adopting isothermal amplification technology
ActiveCN102373301AHigh sensitivityReduce false positive rateMicrobiological testing/measurementMicroorganism based processesPorcine transmissible gastroenteritis virusAfter treatment
The invention relates to the field of molecular-biological detection methods and detection reagents of animal epidemics, in particular to a primer, a kit and a detection method for quickly detecting a porcine transmissible gastroenteritis virus by adopting an isothermal amplification technology (NASBA (Nucleic Acid Sequence-based Amplification)). In the invention, gene sequences of the porcine transmissible gastroenteritis virus are subjected to multiple alignments through ClustalW; conserved areas of the sequences are analyzed; primers and probes are respectively designed by primer 5.0 design software; and the porcine transmissible gastroenteritis virus can be quickly and specifically detected and distinguished. Various reaction solutions are prepared to form a corresponding assay kit. The kit provided by the invention has the advantages of simplicity and convenience for operation, high specificity, high sensitivity, high repeatability, no complex after treatment, wide applicability and the like.
Owner:重庆海关技术中心 +1
NASBA-ELISA (nucleic acid sequence-based amplification-enzyme-linked immuno sorbent assay) detection primer, probe and kit for type II grass carp reovirus
InactiveCN104388588AShort detection time periodImprove detection efficiencyMicrobiological testing/measurementMicroorganism based processesPositive controlNucleotide
The invention discloses an NASBA-ELISA (nucleic acid sequence-based amplification-enzyme-linked immuno sorbent assay) detection primer, a probe and a kit for type II grass carp reovirus. Nucleotide sequences of the primer and the probe are as shown in SEQ ID NO.1-4. The kit contains the primer, the probe, an NASBA amplification buffer solution, an enzyme mixed liquor, an amplification product detection reagent, a negative control and a positive control. The kit for the type II grass carp reovirus has the advantages that a detection time period is short, and detection efficiency is high; virus detection specificity is high, and accuracy is high; virus quantitative analysis can be realized while virus qualitative analysis is realized; detection sensitivity is higher than that of common PCR (polymerase chain reaction); operation is easy, and popularization is easy; and repeatability of experimental results is good.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
Astrovirus real-time isothermal amplification detection kit, its primers and probe
InactiveCN102965453AAvoid inconvenienceStrong specificityMicrobiological testing/measurementMicroorganism based processesDiseaseAvastrovirus
The invention relates to an enteropathogen rapid detection technology based on real-time nucleic acid sequence-based amplification (NASBA). Specifically, the invention provides an Astrovirus real-time isothermal amplification detection kit, and a pair of primers and a molecular beacon probe thereof. The kit includes: a 2*real-time NASBA reaction solution containing the primers and the probe, a 5*enzyme mixed solution and a positive control template, a negative control and a blank control. The sequences of the primers and the probe are the sequences numbered as SEQIDNO:1-3, and the primers and the probe can specifically amplify and detect an Astrovirus ORF2 gene fragment. The kit provided in the invention has the characteristics of fastness, high efficiency, sensitivity and specificity, and real-time detection analysis, etc., and can be used in the fields of conventional detection and disease control and prevention in clinical practice and ports.
Owner:珠海国际旅行卫生保健中心
Virus nucleic acid extraction or preservation reagent, primer probe combination, virus amplification reagent, kit and application thereof
ActiveCN112266986AHigh recovery rateShort timeMicrobiological testing/measurementMicroorganism based processesReference genesViral nucleic acid
The invention relates to the field of virus detection, and particularly relates to a virus nucleic acid extraction or preservation reagent, a primer probe combination, a virus amplification reagent, akit and application thereof. A virus detection device provides a simple and feasible virus nucleic acid extraction method, the whole process is about 5-15 minutes, and purified nucleic acid is recovered and can be used for detecting virus nucleic acid. The method comprises PCR, NASBA, LAMP, RPA and the like. Compared with a traditional virus extraction method, the method is high in virus nucleicacid recovery rate, short in time, convenient to operate and easy to clinically popularize. The invention relates to an isothermal amplification primer, a probe combination sequence and a reaction buffer solution for simultaneously detecting novel coronavirus COVID-19N and ORF genes and human-derived reference genes through a single tube. The system is good in specificity, high in sensitivity (50cp / mL) and high in specificity, only needs 20min of detection time, and can report positive within about 10 min at the soonest.
Owner:CAPITALBIO CORP +1
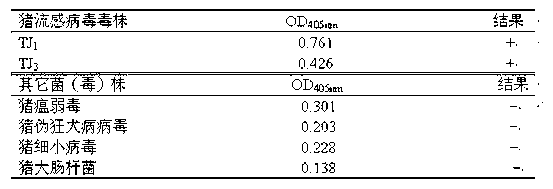
Nucleic-acid sequence-based amplification (NASBA) method for detecting swine influenza virus (SIV)
InactiveCN102534052AMicrobiological testing/measurementMicroorganism based processesQuarantineSwine influenzavirus
The invention discloses a nucleic-acid sequence-based amplification (NASBA) method for detecting a swine influenza virus (SIV). The NASBA method mainly comprises the following steps of: extracting the ribonucleic acid (RNA) from the SIV, preparing an NASBA system, preparing an NASBA program, carrying out identification on an NASBA product and carrying out PCR (Polymerase Chain Reaction) amplification. According to the NASBA method, the NASBA rapid detection method is established by taking the NP gene of the SIV as a target gene and is used for the diagnosis of the SIV. The method has higher specificity and sensitivity so as to detect a virus solution with the dilution factor of 10<-5>, so that the method can be used for the rapid detection of clinically-suspected SIV samples, meanwhile, the technical level of China in the diagnosis, epidemic surveillance, inspection and quarantine of swine influenza can be improved so as to ensure the healthful and rapid development of swine industry.
Owner:天津市宁河原种猪场有限责任公司
Swine fever virus detection method based on G-quadruplex fluorescence characteristic and kit
InactiveCN104611462AHigh amplification efficiencyRapid diagnosisMicrobiological testing/measurementMicroorganism based processesClassical swine fever virus CSFVSwine Fever Virus
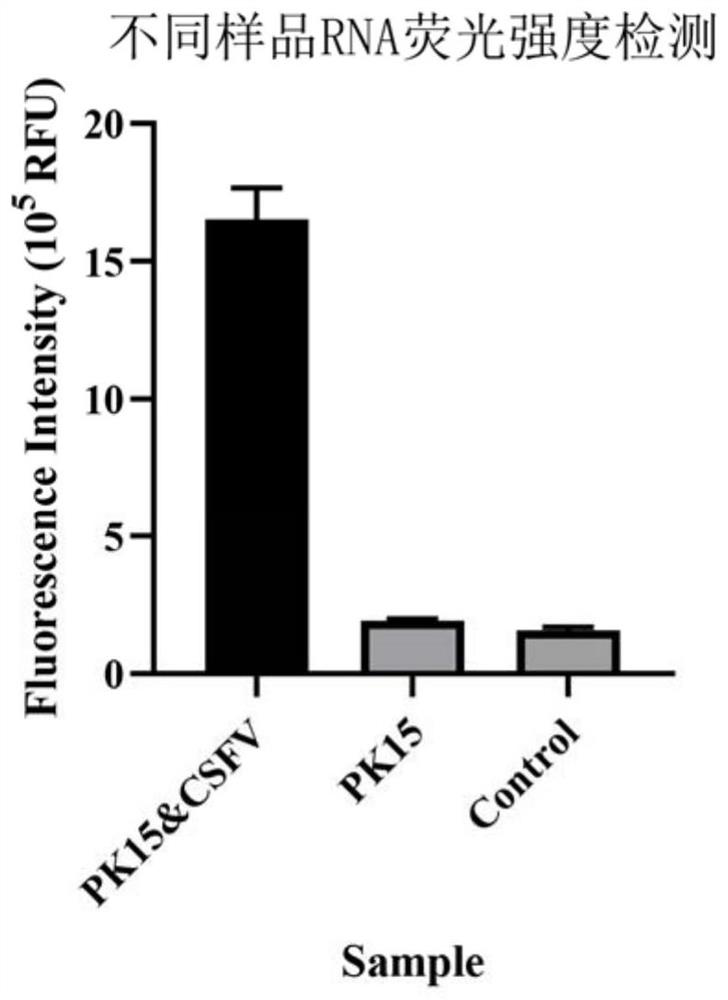
The invention discloses a swine fever virus detection method based on G-quadruplex fluorescence characteristic and a kit, and belongs to the technical field of gene engineering. The method comprises: utilizing a nucleic acid sequence-based amplification (NASBA) technology to amplify target RNA to obtain a single-chain RNA product; adding an upstream probe, a downstream probe and the RNA amplification product into a G- quadruplex fluorescence detection buffer, performing denaturation and annealing, then adding protoporphyrin, and utilizing a microplate reader or fluorophotometer to detect the fluorescence intensity in the reaction system, so as to determine whether the sample possesses a swine fever virus. The method is fast, precise and stable in swine fever virus detection speed, good in reappearance, simple in detection steps and low in cost.
Owner:GUIZHOU DAXING AGRI SCI & TECH DEV CO LTD +1
NASBA method for detecting tomato spotted wilt virus
InactiveCN104404173AReduce the number of cyclesHigh precisionMicrobiological testing/measurementMicroorganism based processesDisease monitoringTomato spotted wilt virus
The invention provides an NASBA method for detecting a tomato spotted wilt virus (TSWV). The sequences of NASBA primers for detecting the TSWV are SEQ ID No: 1-2 respectively. According to the highly conserved region of gene N of the TSWV, two inner primers with specificity are designed. The conserved gene sequences are shared by different strains with TSWV to ensure the reliability in detecting different sources of TSWV at the level of strains. The NASBA method is suitable for rapid detection and confirmation of TSWV and can be widely used in disease monitoring in production and environment, as well as TSWV confirmation in the import and export trade.
Owner:INSPECTION & QUARANTINE TECH CENT SHANDONG ENTRY EXIT INSPECTION & QUARANTINE BUREAU
Macrobrachium rosenbergii Nodavirus NASBA-LFD detection method and detection kit thereof
ActiveCN103409556AStrong specificityHigh detection sensitivityMicrobiological testing/measurementRNA extractionNucleic acid
The invention discloses a Macrobrachium rosenbergii Nodavirus NASBA-LFD (nuclear acid sequence-based amplification-lateral flow dipstick) detection method and a detection kit thereof. The detection method includes: according to a Macrobrachium rosenbergii Nodavirus (MrNV) sequence published by GenBank, screening a conserved region, and designing the primers and probe needed by an MrNV NASBA-LFD reaction system; employing a tissue RNA extraction reagent prepared by the inventor to extract the RNA of a to-be-detected sample, then utilizing the MrNV NASBA reaction system established by the invention to perform detection, and at the end of the reaction, determining the result according to nucleic acid rapid test paper. Compared with common detection technologies of the virus, the method and the kit provided in the invention have the characteristics of simplicity and rapidity, good specificity and high sensitivity, can be used for field service by technical personnel and farmers at the production front line, and are in favor of diagnosis prevention and pathogen blocking of Macrobrachium rosenbergii white tail disease.
Owner:ZHEJIANG INST OF FRESH WATER FISHERIES
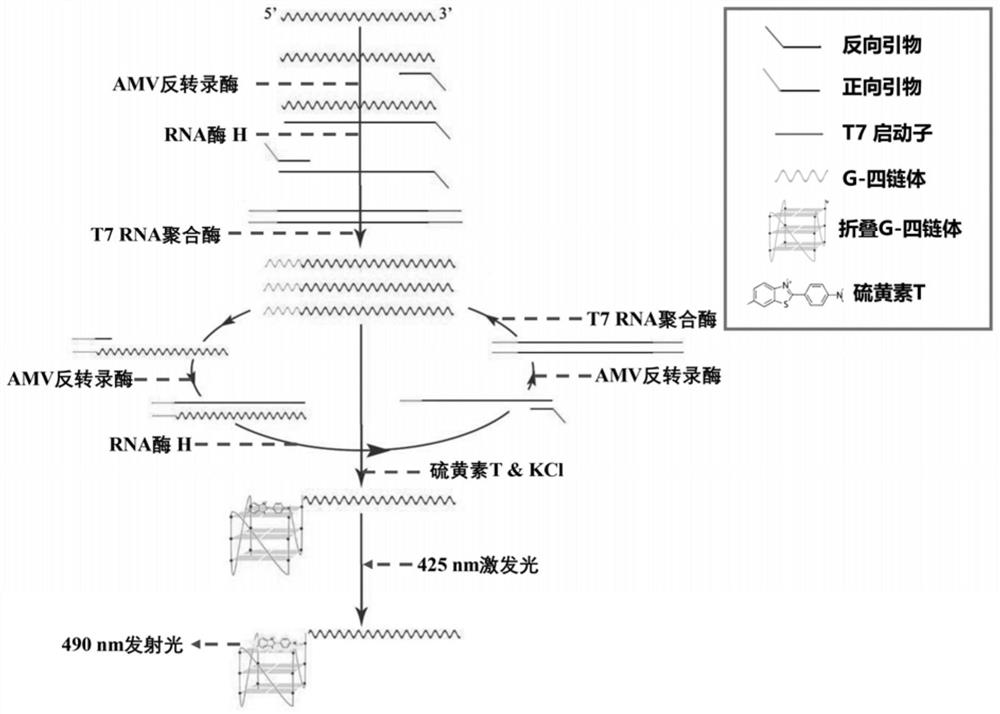
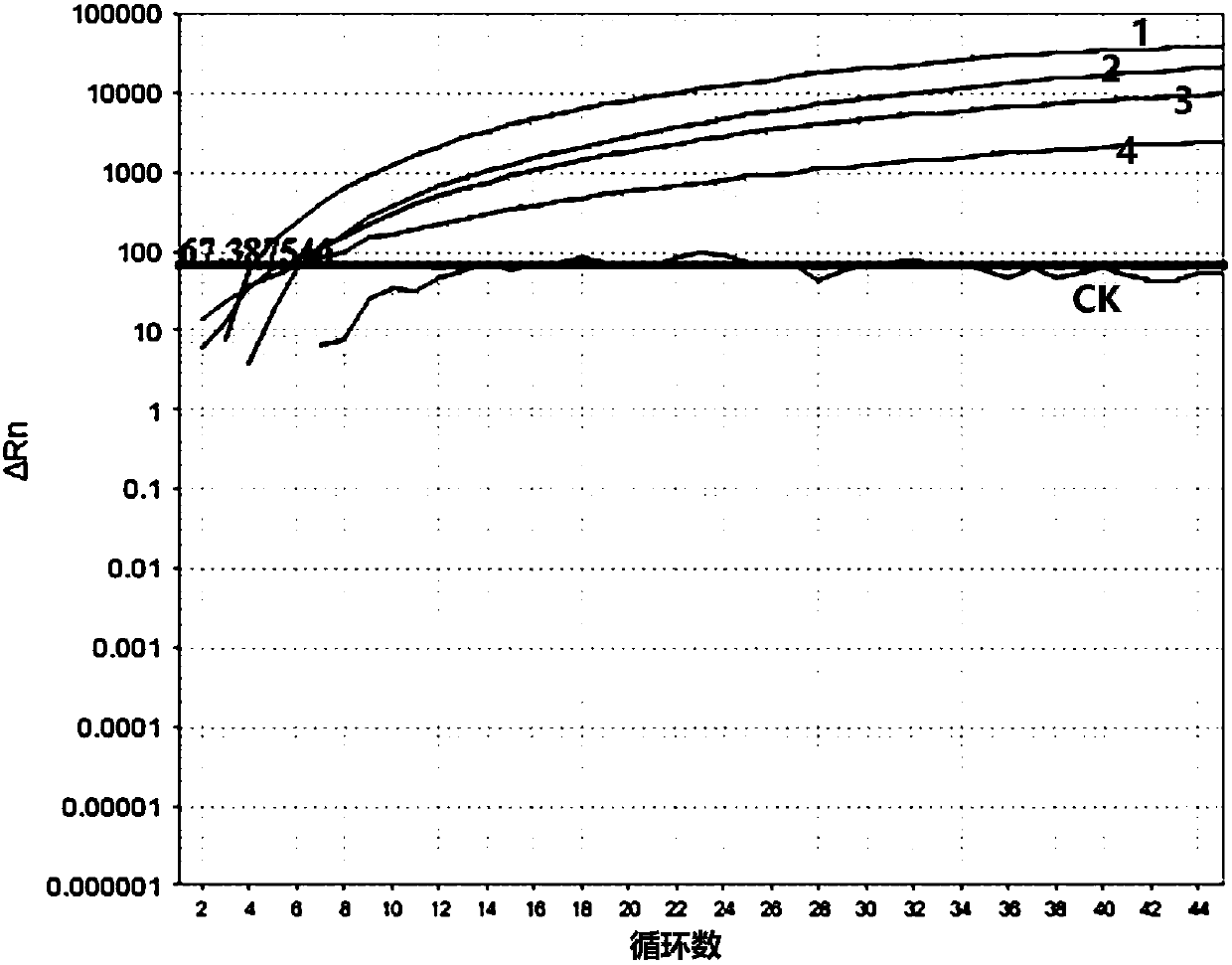
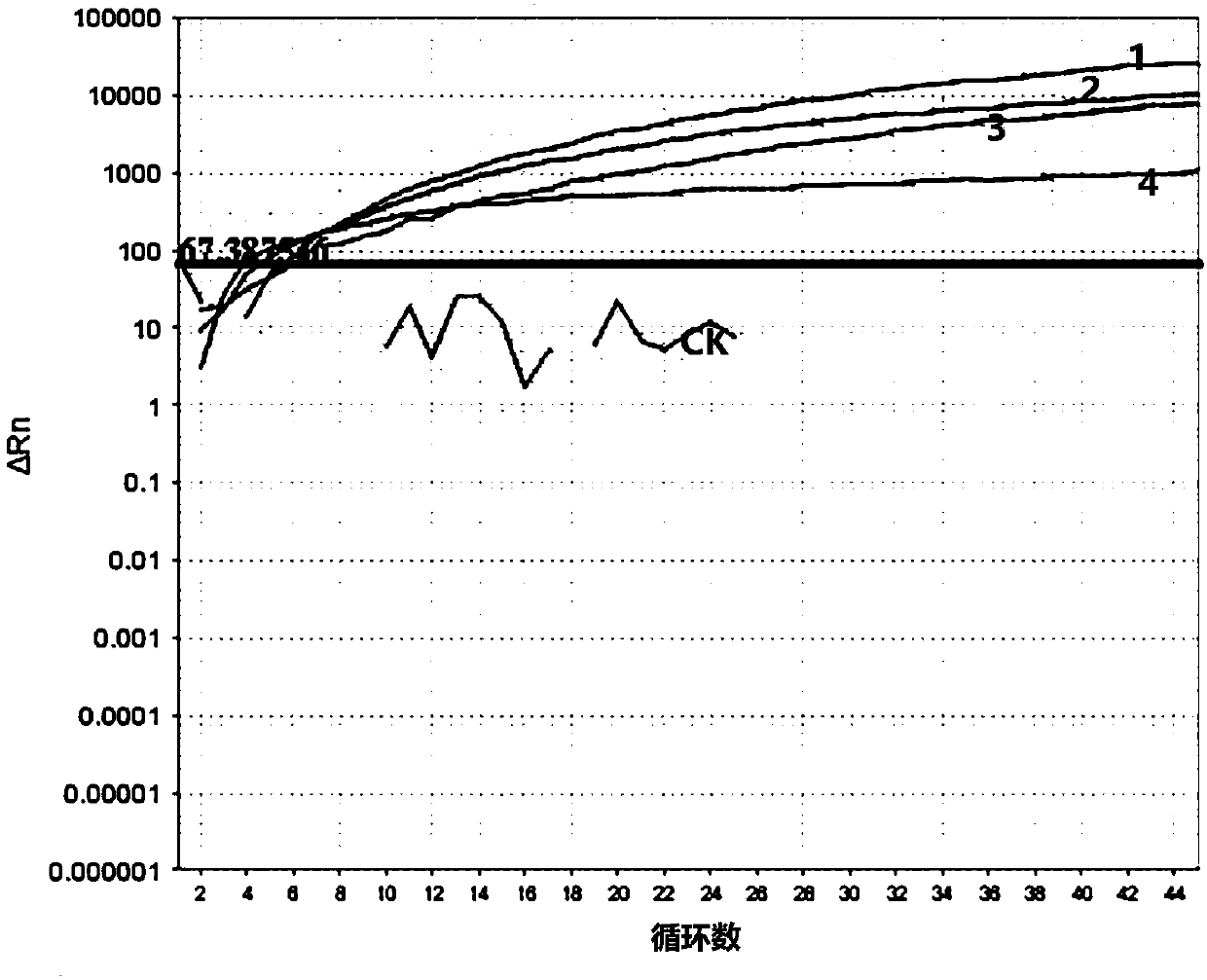
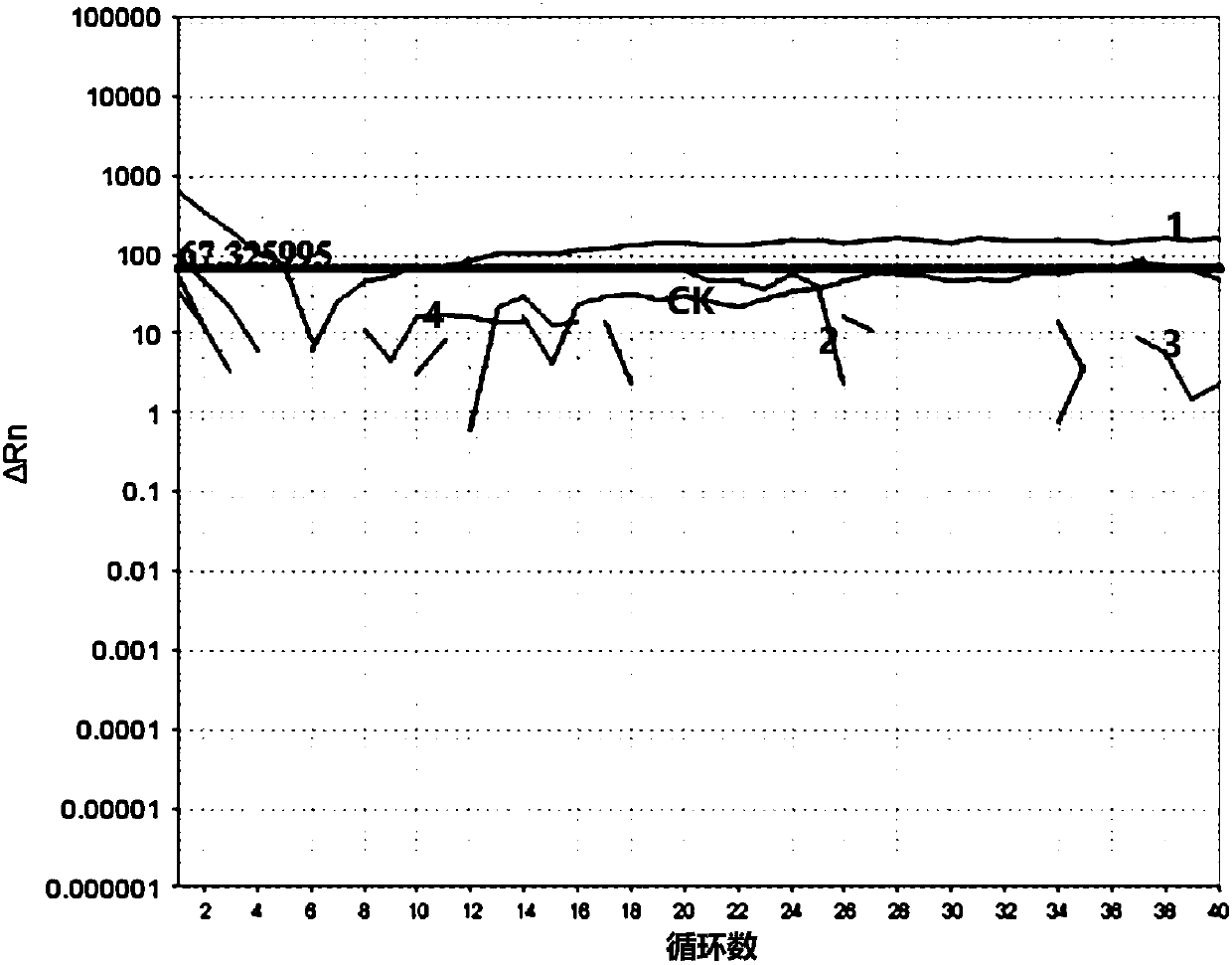
Classical swine fever virus detection method based on G4-ThT biosensor and NASBA and kit thereof
PendingCN114875176AFluorescent response is strong and significantStrong fluorescence responseMicrobiological testing/measurementMicroorganism based processesClassical swine fever virus CSFVThioflavin
The invention belongs to the technical field of gene engineering, and particularly relates to a hog cholera virus detection method and kit based on a G4-ThT biosensor and NASBA, and the specific method comprises the following steps: taking virus RNA extracted from a hog cholera virus sample as an amplification object, carrying out isothermal amplification by adopting an isothermal nucleic acid amplification technology, then adding a buffer solution containing Tris-HCl and KCl, and carrying out isothermal amplification by adopting an isothermal nucleic acid amplification technology; then thioflavin T is added to promote G-quadruplex on an amplification product to be folded and form a compound, fluorescence is detected under the excitation condition of a light source with a specific wavelength, and judgment is carried out according to the intensity of the fluorescence. The invention provides a detection method and a kit which do not need special biotin labeling or sulfydryl modification treatment probes, and the detection cost is obviously reduced.
Owner:SOUTH CENTRAL UNIVERSITY FOR NATIONALITIES
NASBA detection method for identifying 1/2c serotype of listeria monocytogenes
ActiveCN107723348AEasy to distinguishAccurate distinctionMicrobiological testing/measurementDNA/RNA fragmentationForward primerRNA extraction
The invention discloses an NASBA detection primer and an NASBA detection method for identifying the 1 / 2c serotype of listeria monocytogenes, belonging to the field of biotechnology. A forward primer sequence is 5'-GAAATACATTTTGGTCTAG-3', a reverse primer sequence is 5'-aattctaatacgactcactatagggATTACTGGGAATAACTTGA-3', a sequence of a molecular beacon probe for detecting the 1 / 2c serotype of listeria monocytogenes is 5'-JOE-CGATCGAGGAAATGCTTATGCAGATATTACGATCG-DABCYL-3'. The detection method comprises the following steps: extracting RNA of listeria monocytogenes, carrying out a real-time NASBA reaction, and carrying out qualitative analysis. The constructed primer and molecular beacon have high specificity and sensitivity for the identification of the 1 / 2c serotype strain of the listeria monocytogenes, for the detection method, the operation steps are simple, the detection time is short, real-time qualitative analysis can be carried out, and meanwhile, living bacteria and dead bacteria can be effectively distinguished.
Owner:NANJING AGRICULTURAL UNIVERSITY
NASBA kit for detecting monkey pox virus and application thereof
InactiveCN103468830BQuick screeningGuaranteed specificityMicrobiological testing/measurementFluorescence/phosphorescenceSingle strandQuarantine
The invention discloses an NASBA kit for detecting a monkey pox virus and the application thereof. The NASBA kit contains a composition used for detecting the monkey pox virus, the composition is composed of a primer pair and a probe, the primer pair is composed of two single-stranded DNA molecules shown in the first sequence and the second sequence in a sequence list, and the sequence of the probe is the third sequence in the sequence list. The NASBA kit is used for detecting the monkey pox virus, and the advantages of being simple, convenient, efficient, quick and peculiar are achieved. The NASBA kit can quickly screen the monkey pox virus and has great significance to screening port input diseases, technical support and strategic reserve are well prepared for preventing introduction of the diseases, and the demand of current health quarantine is met.
Owner:浙江国际旅行卫生保健中心
NASBA primer, kit and method for detecting peach latent mosaic viroid
InactiveCN104498633AReduce the number of cyclesHigh precisionMicrobiological testing/measurementMicroorganism based processesBiotechnologyElectrophoreses
The invention discloses an NASBA primer, a kit and a method for detecting peach latent mosaic viroid. The sequences of an upstream primer and a downstream primer are respectively SEQ ID No. 1-2. An NASBA amplification kit comprises the following components: (1) NASBA amplification reaction liquid A; (2) NASBA amplification reaction liquid B. The detection method is as follows: 1) extracting sample RNA; 2) carrying out PLMVd NASBA amplification; and 3) carrying out electrophoresis detection. The NASBA primer, kit and method disclosed by the invention are suitable for quick detection and verification of peach latent mosaic viroid and can be widely used for monitoring epidemic situations in agricultural production and environment and monitoring and detecting the kind of viruses in import and export trades, the operation is very simple and convenient, the necessary sample amount is small and the requirements on the quality of template RNA are quite low.
Owner:INSPECTION & QUARANTINE TECH CENT SHANDONG ENTRY EXIT INSPECTION & QUARANTINE BUREAU
Method of identifying severe acute respiratory syndrome corona virus 2 (sars-cov-2) ribonucleic acid (RNA)
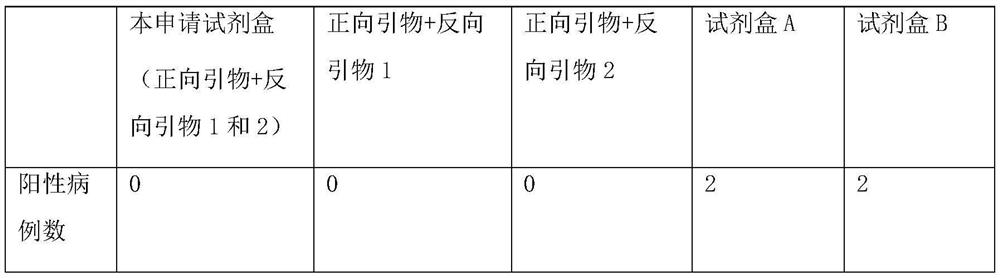
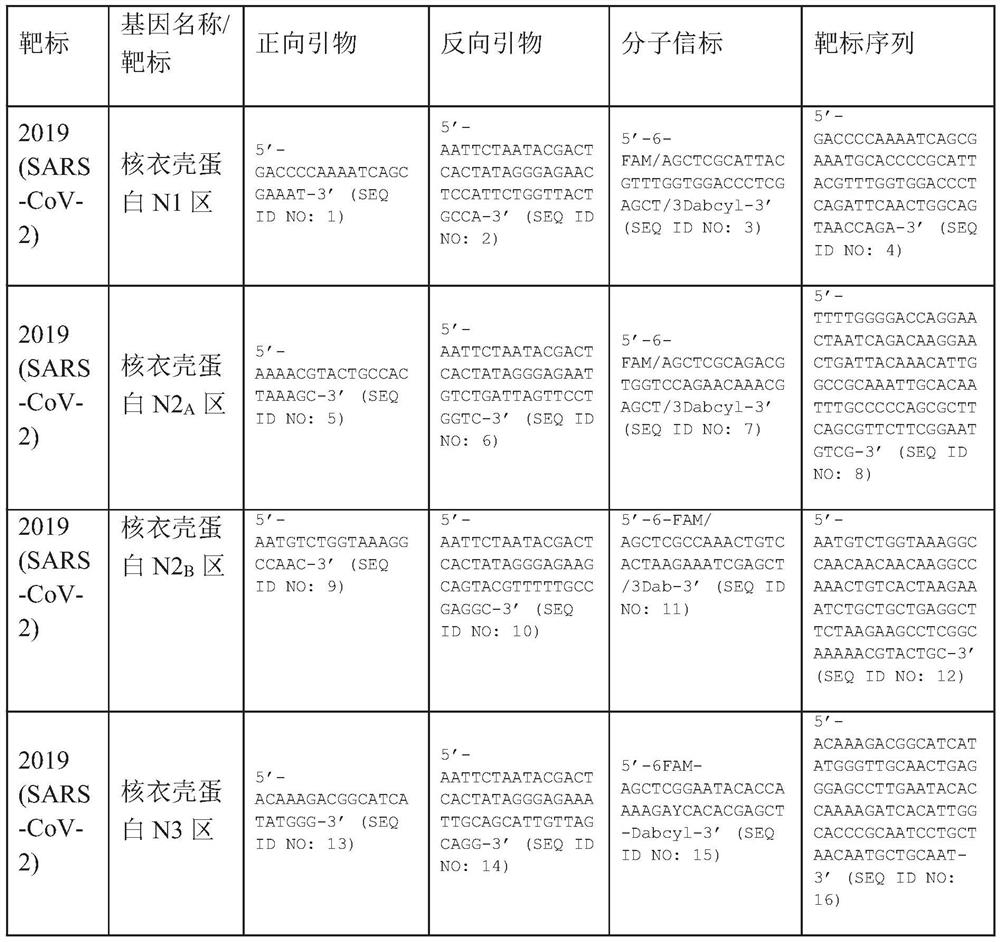
A method for detecting the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a biological sample includes the steps of lysing the biological sample to form a lysate and generating an amplified lysate by performing a nucleic acid sequence-based (NASBA) amplification for a target nucleic acid sequence in the lysate in the presence of: a forward primer having the oligonucleotide sequence selected from the group consisting of SEQ ID NO: 1, SEQ ID NO: 5, SEQ ID NO: 9, SEQ ID NO: 13, and SEQ ID NO: 17; a reverse primer having the oligonucleotide sequence selected from the group consisting of SEQ ID NO: 2, SEQ ID NO: 6, SEQ ID NO: 10, SEQ ID NO: 14, and SEQ ID NO: 18; and a molecular beacon having the oligonucleotide sequence selected from the group consisting of SEQ ID NO: 3, SEQ ID NO: 7, SEQ ID NO: 11, SEQ ID NO: 15, and SEQ ID NO: 19 and a fluorophore. The amplified lysate is exposed to an excitation source. A fluorescence of the fluorophore is detected in the amplified lysate exposed to the excitation source The SARS-CoV-2 is determined to be present in the biological sample in response to detecting the fluorescence of the fluorophore.
Owner:TELEFLEX MEDICAL INC
Method for detecting REV by using NASBA, and reagent set used therein
ActiveCN113025746AStrong specificityHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationVirusDNA
Owner:BOAO BIOLOGICAL CO LTD
2019-nCoV nucleic acid isothermal amplification detection kit based on SYBR Green I and detection method
PendingCN111826470AQuality improvementHigh quality and high concentrationMicrobiological testing/measurementAgainst vector-borne diseasesVirusBioinformatics
The invention discloses a 2019-nCoV nucleic acid isothermal amplification detection kit based on SYBR Green I and a detection method. The kit comprises an ORF1ab gene primer sequence and an N gene primer sequence of a 2019-nCoV virus, an SYBR Green I dye, an NASBA amplification buffer solution and an NASBA enzyme mixed solution. The detection method using the kit comprises the following steps: firstly, preparing the NASBA amplification buffer solution, preparing the NASBA enzyme mixed solution, preparing a primer mixed solution and preparing an RNA sample; and carrying out mixed reaction on all the solutions, carrying out incubating, and finally completing detection by using a fluorescence detector. The kit and the method show high-specificity and high-sensitivity detection performance, and a large amount of experimental time is saved.
Owner:HANGZHOU YORK BIOTECH CO LTD
Nasba primers, kits and methods for detecting apple rust viroids
InactiveCN104450972BGuaranteed reliabilityReduce the number of cyclesMicrobiological testing/measurementMicroorganism based processesElectrophoresisReagent
The invention discloses a NASBA primer, a reagent kit, and a method for detecting apple scar skin viroid; the sequences of the upstream primer and the downstream primer are respectively SEQ ID NO: 1-2; the NASBA augmentation reagent kit comprises the constituents as follows: (1) NASBA augmentation reaction liquid A; (2) NASBA augmentation reaction liquid B; the detection method is as follows: 1) extracting sample RNA; 2) augmenting NASBA of ASSVd; 3) detecting with electrophoresis. The NASBA primer, the reagent kit, and the method for detecting apple scar skin viroid are applicable to quickly detecting and confirming apple scar skin viroid, can be widely applied for epidemic surveillance in agricultural production and environment as well as monitoring and detecting the viroid in import and export trade; the operation is very simple; the amount of the required sample is small; and the quality requirements to template RNA is lower.
Owner:INSPECTION & QUARANTINE TECH CENT SHANDONG ENTRY EXIT INSPECTION & QUARANTINE BUREAU
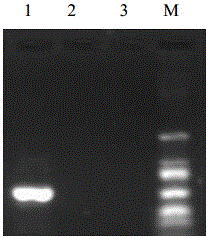
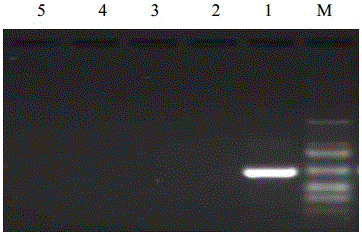
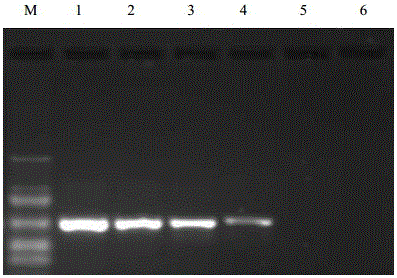
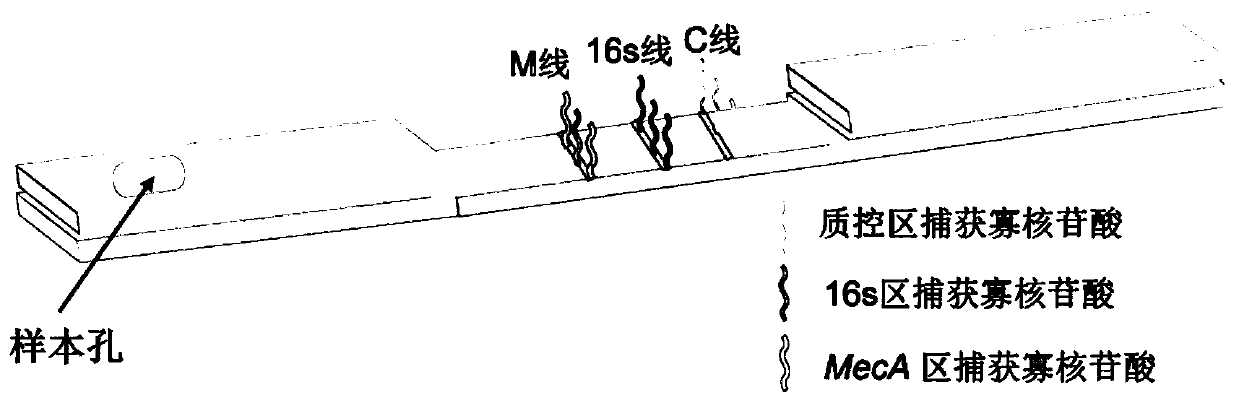
Kit and method for rapid detection of staphylococcus meca
ActiveCN110029182BTest results are stableHigh sensitivityMicrobiological testing/measurementMicroorganism based processesReagent stripMethicillin resistant Staphylococcus
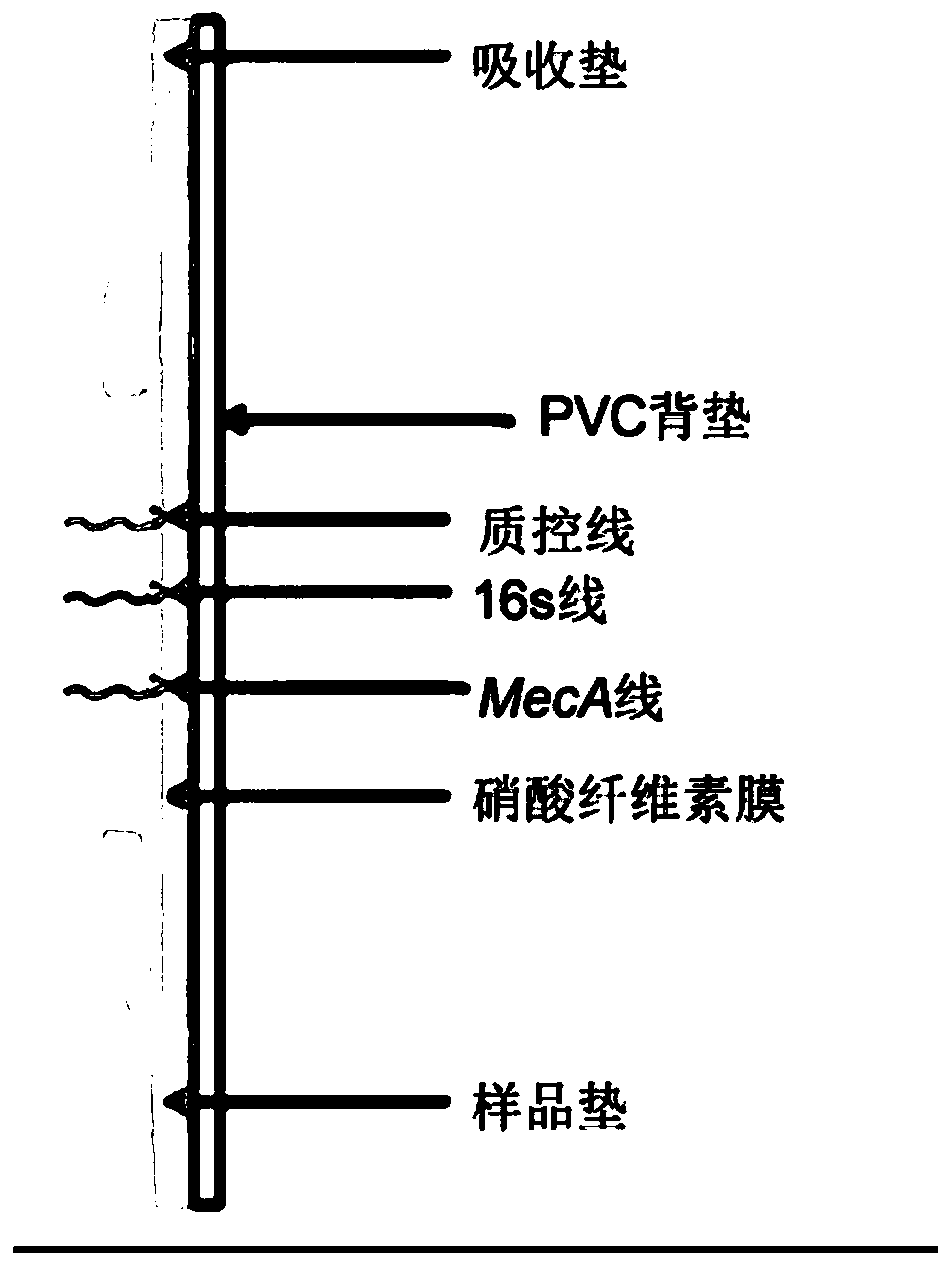
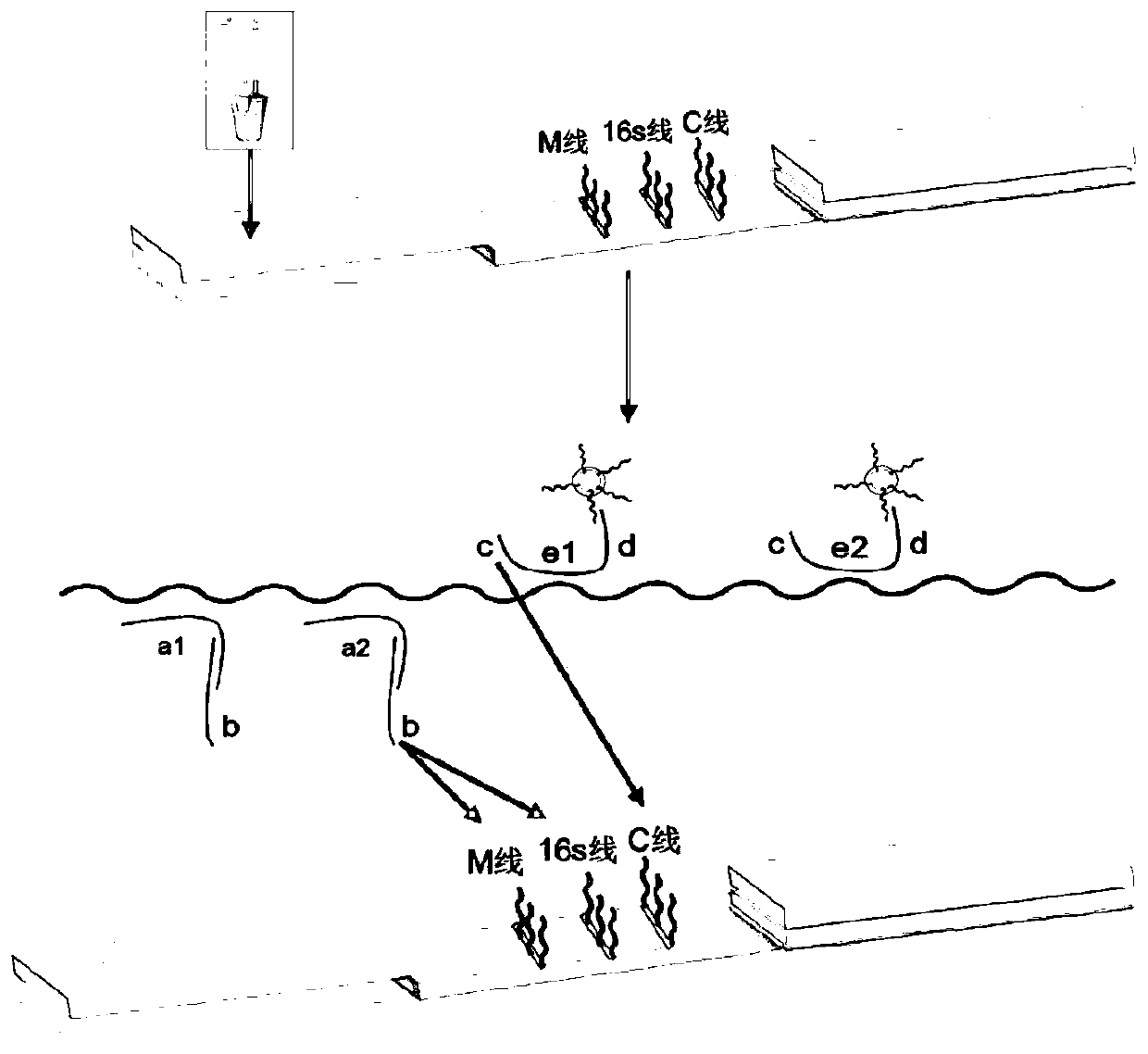
The invention discloses a reagent kit and method for quickly detecting staphylococcus MecA. The reagent kit comprises cell lysate, amplifying reaction liquid, colloidal gold detection liquid and an immunochromatography reagent strip. The reagent kit is characterized in that transcription products of MecA and 16S rRNA are obtained through NASBA amplification on a primer pair in the amplifying reaction liquid. The reagent kit for quickly detecting genes of the staphylococcus MecA is stable in detection result, high in accuracy rate, high in sensitivity, low in cost and short in time consumptionand suitable for clinical promotion. The detection method is suitable for clinically and quickly detecting methicillin resistant staphylococcus and screening staphylococcus at pharynx nasalis, is quick and accurate, is simple in equipment, is low in requirements for operators, and has potential in bedside detection and application in hospitals at different levels.
Owner:WUHAN UNIV
Kit and detection method for quickly detecting porcine transmissible gastroenteritis virus by adopting isothermal amplification technology
ActiveCN102373301BRapid identificationRapid differential testMicrobiological testing/measurementMicroorganism based processesAssayNucleic acid sequencing
Owner:重庆海关技术中心 +1
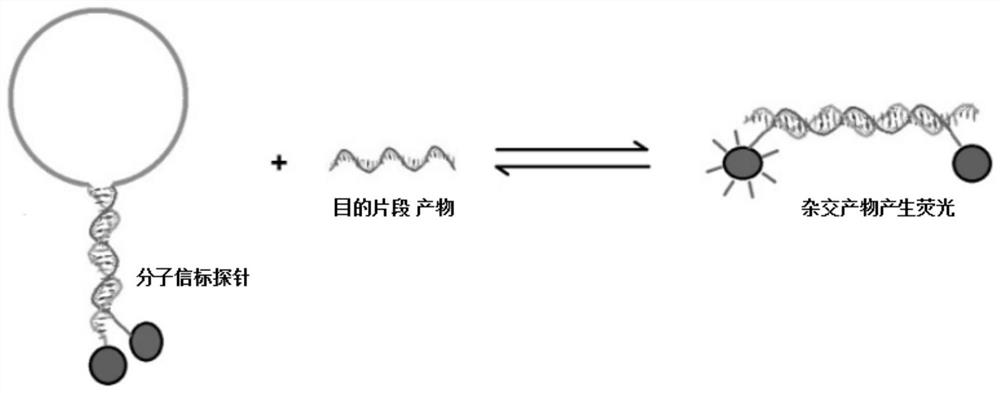
2019-nCoV nucleic acid isothermal amplification primer based on molecular beacon, molecular beacon, test kit and detection method
PendingCN111826469AQuality improvementHigh quality and high concentrationMicrobiological testing/measurementAgainst vector-borne diseasesVirusBioinformatics
The invention discloses a 2019-nCoV nucleic acid isothermal amplification primer based on a molecular beacon, the molecular beacon, a test kit and a detection method. The 2019-nCoV virus primer comprises an ORF1ab gene primer sequence and an N gene primer sequence of a 2019-nCoV virus; the molecular beacon comprises an ORF1ab gene molecular beacon probe and an N gene molecular beacon probe; the test kit comprises the 2019-nCoV virus primer, a molecular beacon, an NASBA amplification buffer solution and an NASBA enzyme mixed solution, the detection method using the test kit comprises the following steps: firstly, preparing an NASBA amplification buffer solution, preparing an NASBA enzyme mixed solution, preparing a primer mixed solution, and preparing an RNA sample; mixing, reacting and incubating all the solutions, and finally completing detection through a fluorescence detector. The invention shows high-specificity and high-sensitivity detection performance, and a large amount of experiment time is saved.
Owner:HANGZHOU YORK BIOTECH CO LTD
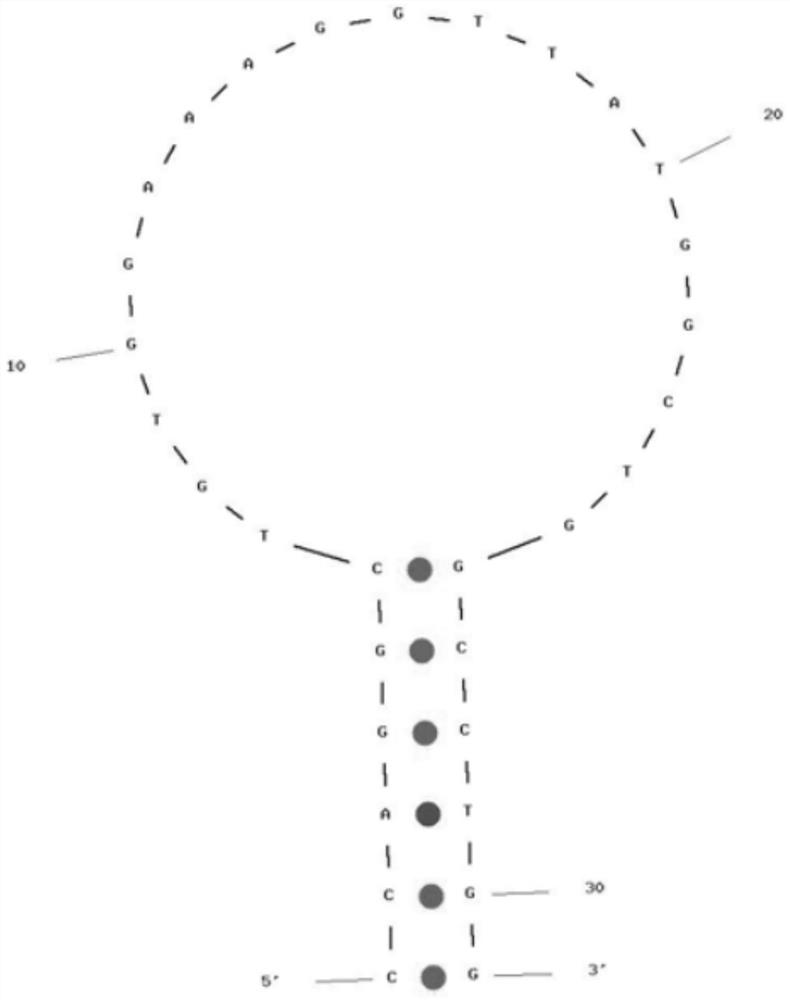
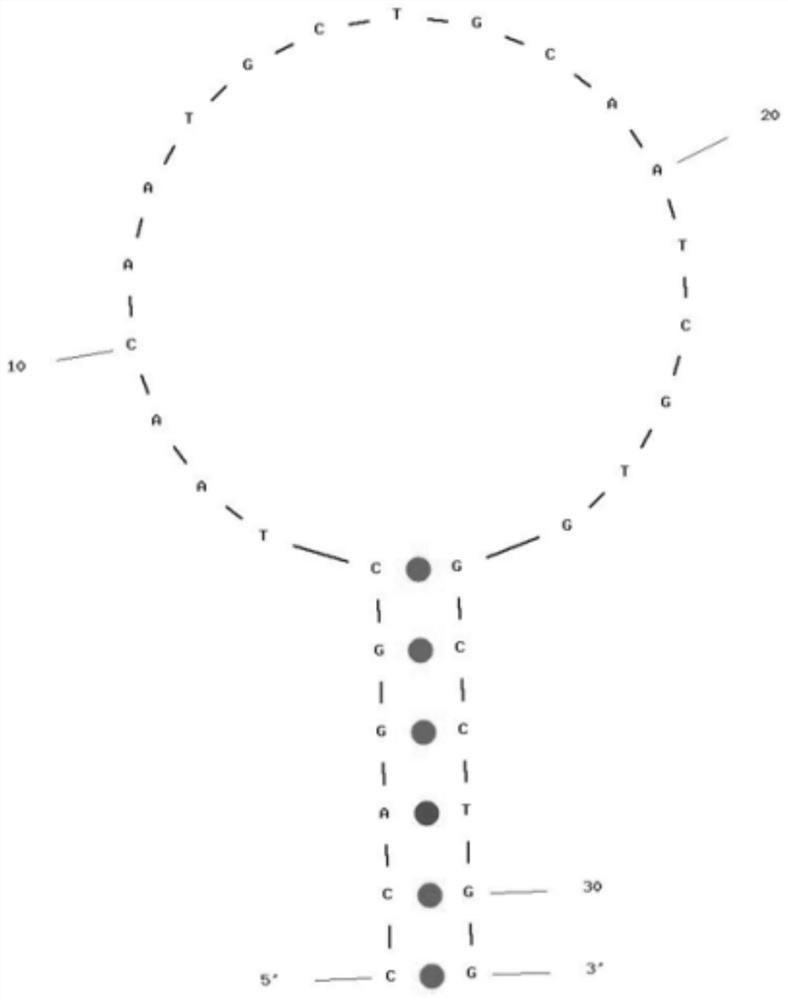
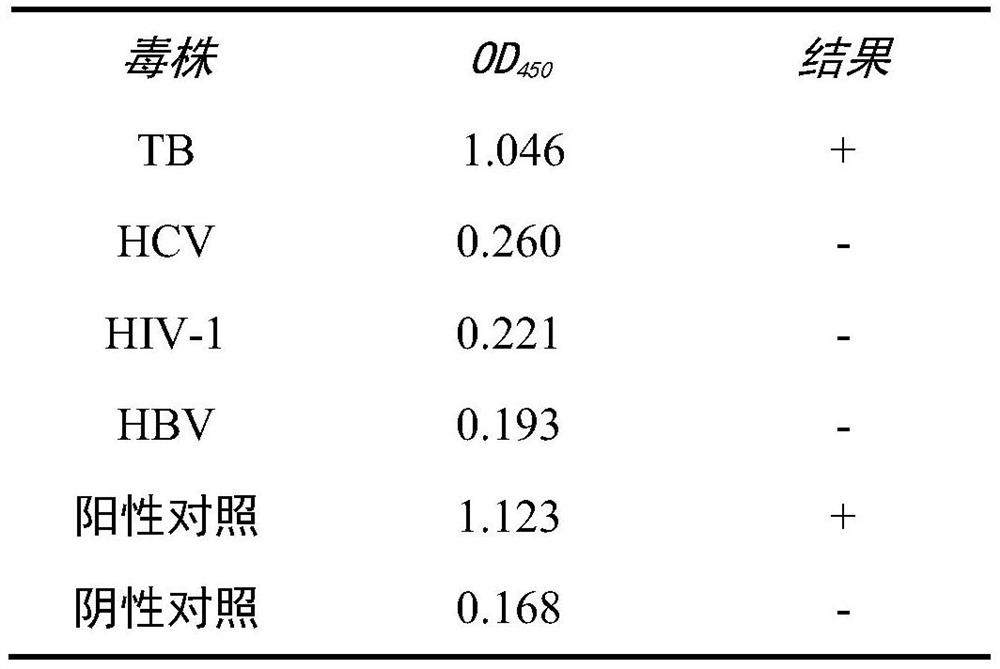
A rapid detection kit and detection method for mycobacterium tuberculosis nasba-elisa in deer
ActiveCN106755546BEasy to operateAvoid high temperature denaturation stepsMicrobiological testing/measurementTuberculosis mycobacteriumMycobacterium
The invention relates to the technical field of biological detection, and is a rapid detection kit for Mycobacterium tuberculosis NASBA-ELISA in deer, which is characterized in that it consists of specific primers and capture probes designed according to the sequence of the conserved region of Mycobacterium tuberculosis 16srRNA gene. needle, detection probe, optimized NASBA amplification system and optimized NASBA product detection system. It can perform simple and rapid detection of Mycobacterium tuberculosis in deer, has the advantages of high specificity and easy promotion, and is also helpful for the early diagnosis of bovine and human multiple Mycobacterium tuberculosis; the detection method is designed and artificially Synthesize specific primers, capture probes and detection probes for the conserved region of the Mycobacterium tuberculosis gene sequence, determine the steps of the NASBA amplification reaction program and the NASBA product detection system, and can simultaneously monitor human and bovine multiple Mycobacterium tuberculosis, The detection result has high sensitivity and good repeatability.
Owner:JILIN AGRI SCI & TECH COLLEGE
Viral nucleic acid extraction or preservation reagents, primer probe combinations, virus amplification reagents, kits and their applications
ActiveCN112266986BHigh recovery rateShort timeMicrobiological testing/measurementMicroorganism based processesReference genesViral nucleic acid
Owner:CAPITALBIO CORP +1
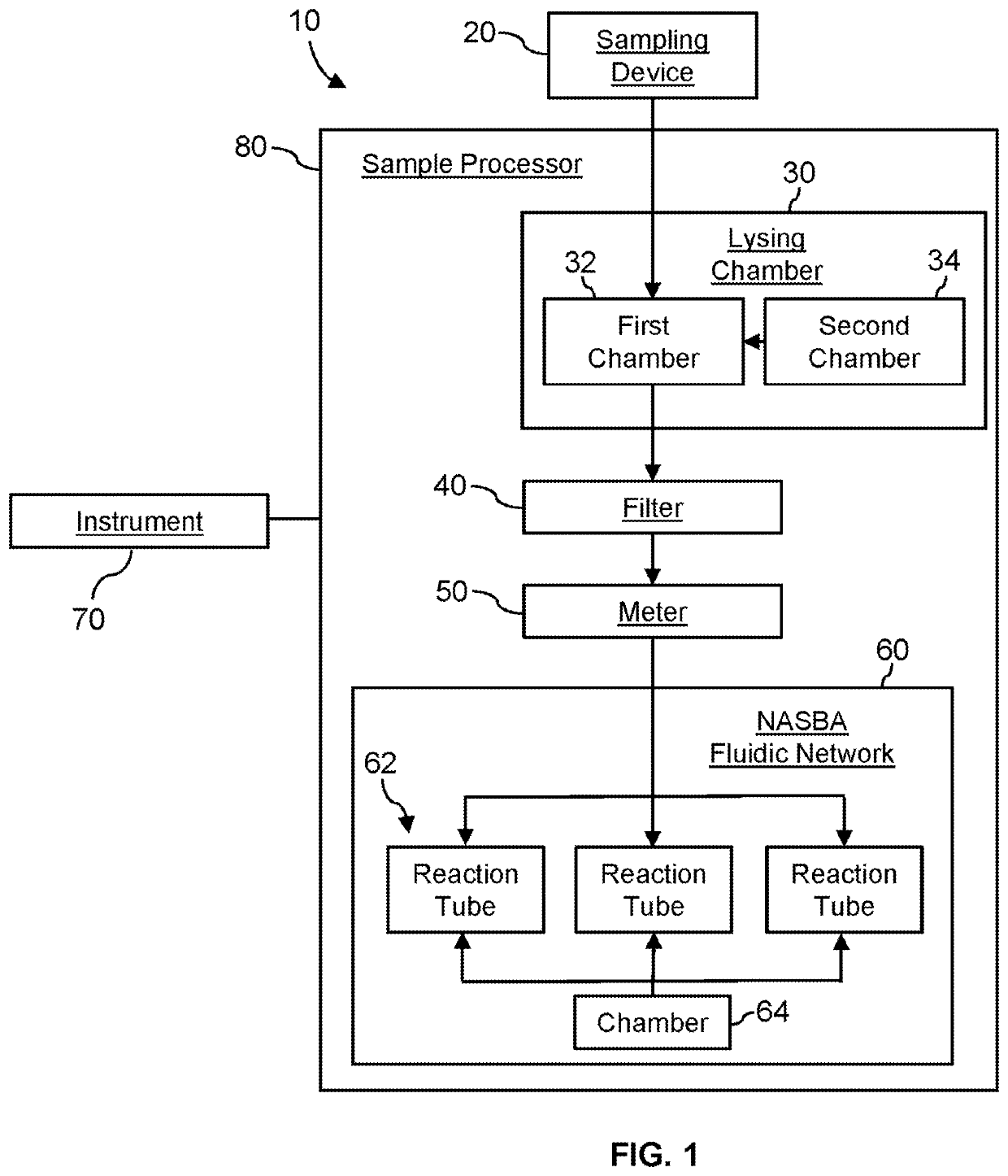
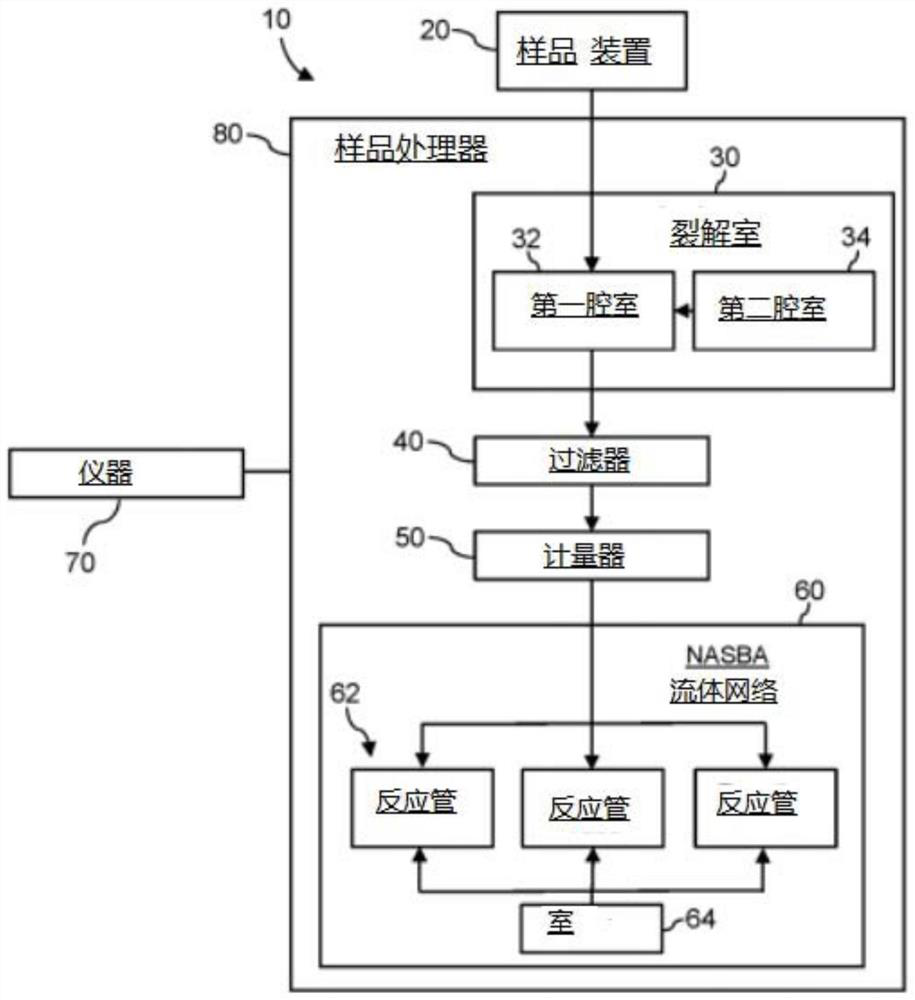
System for identifying severe acute respiratory syndrome corona virus 2 (sars-cov-2) ribonucleic acid (RNA)
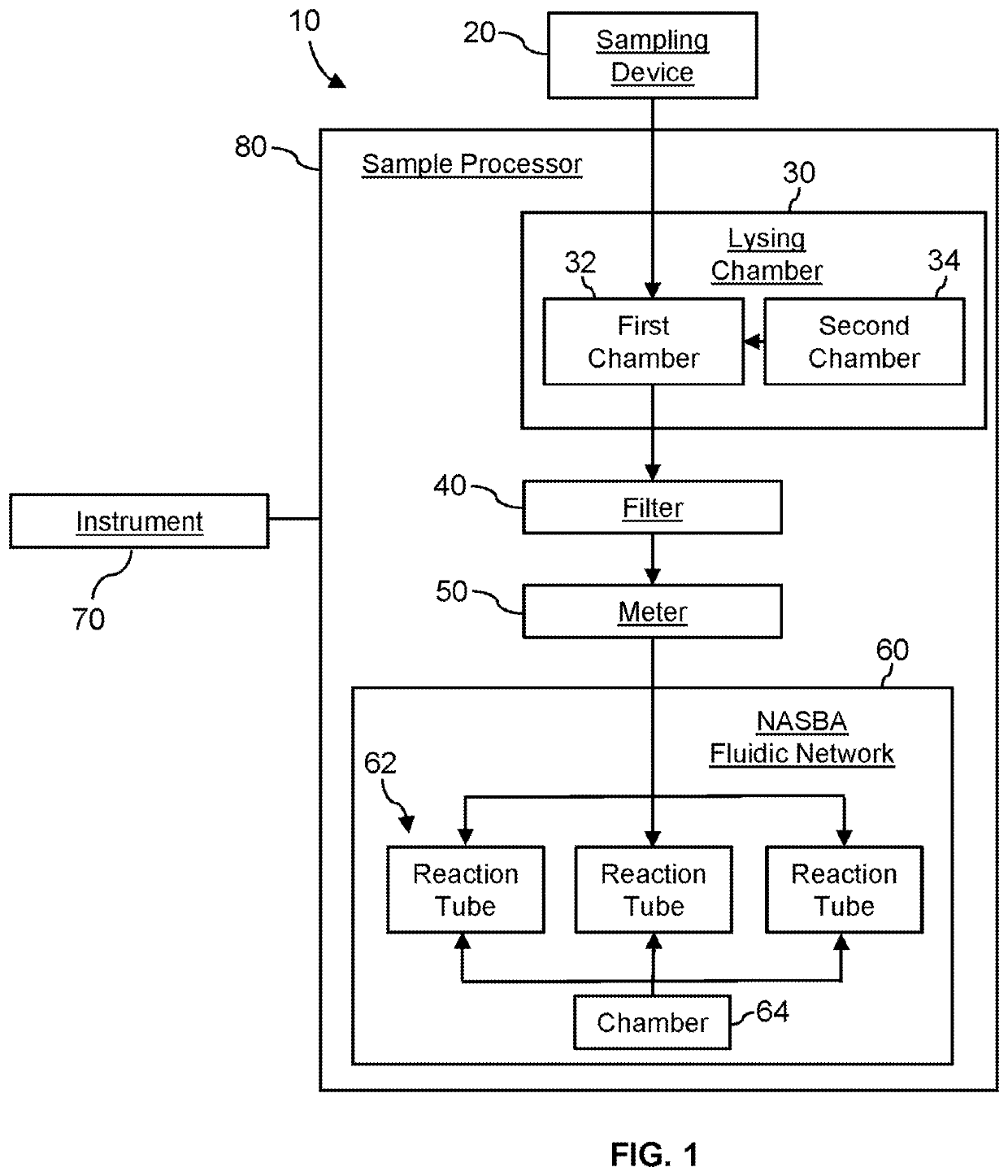
InactiveUS20210129144A1Microbiological testing/measurementLaboratory glasswaresForward primerFluorophore
A system for detecting the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a biological sample includes a sampling device, a lysing chamber, a NASBA fluidic network, and an analytical instrument. The sampling device is configured to contain a sample containing a pathogen target sequence for SARS-CoV-2. The lysing chamber is configured to be in fluid communication with the sampling device to receive the sample. The is lysing chamber is configured to lyse the sample into a lysate. The NASBA fluidic network is configured to be in fluid communication with the lysing chamber to receive the lysate. The NASBA fluidic network includes an enzyme, a forward primer, and a reverse primer for amplifying a predetermined genetic sequence in the pathogen target sequence contained within the lysate. The forward primer has the oligonucleotide sequence selected from the group consisting of SEQ ID NO: 1, SEQ ID NO: 5, SEQ ID NO: 9, SEQ ID NO: 13, and SEQ ID NO: 17. The reverse primer has the oligonucleotide sequence selected from the group consisting of SEQ ID NO: 2, SEQ ID NO: 6, SEQ ID NO: 10, SEQ ID NO: 14, and SEQ ID NO: 18. A molecular beacon is configured to attach to the pathogen target sequence. The beacon has the oligonucleotide sequence selected from the group consisting of SEQ ID NO: 3, SEQ ID NO: 7, SEQ ID NO: 11, SEQ ID NO: 15, and SEQ ID NO: 19 and a fluorophore. The analytical instrument is configured to excite the beacon when the molecular beacon is attached to the pathogen target sequence to signal a presence of the pathogen target sequence.
Owner:TELEFLEX MEDICAL INC
Nucleic acid sequence-based amplification-enzyme-linked immunosorbent assay (NASBA-ELISA) kit for type A swine influenza viruses
ActiveCN102586488BSimple and fast operationHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationAssayNucleic acid sequencing
The invention relates to a kit for a nucleocapsid protein (NP) gene of type A swine influenza viruses. The kit consists of a pair of amplification primers, a capture probe, a detection probe, a nucleic acid sequence-based amplification (NASBA) amplification reagent, and an NASBA product detection reagent. An NASBA rapid detection method is established by taking the NP gene of the type A swine influenza viruses as a target gene, and is used for diagnosing the type A swine influenza viruses, so that the technical levels of the diagnosis, epidemic monitoring, and inspection and quarantine of swine influenza in China are improved, and the healthy and rapid development of pig husbandry is guaranteed.
Owner:天津市农业科学院
A kind of influenza A virus detection primer, probe and kit thereof
ActiveCN111004867BQuick checkAccurate detectionMicrobiological testing/measurementDNA/RNA fragmentationPcr methodInfluenza Viruses Type A
The invention relates to an influenza A virus detection kit and application thereof. The kit provided by the invention contains a primer group and a probe for influenza virus detection, and specifically comprises three primers and one probe, and is a primer and probe combination designed specific for influenza A virus NP target gene. The primers and the probe disclosed by the invention are suitable for detecting influenza A virus by NASBA method, support high-throughput, rapid and accurate detection of influenza virus infection, can acquire more accurate detection result within one hour than real-time fluorescent quantitative PCR method, and have important significance for clinical application.
Owner:MUDANJIANG MEDICAL UNIV
Detection system for severe acute respiratory syndrome coronavirus 2
The invention relates to a detection system for severe acute respiratory syndrome coronavirus 2. The detection system comprises a sampling device, a cracking chamber, an NASBA fluid network and an analysis instrument. The sampling device is configured to contain a sample containing a pathogen target sequence for SARS-CoV-2. The NASBA fluid network includes an enzyme, a forward primer and a reverse primer for amplifying a predetermined gene sequence in a pathogen target sequence contained within the lysate. The forward primer has an oligonucleotide sequence selected from the following groups: SEQ ID NO: 1, SEQ ID NO: 5, SEQ ID NO: 9, SEQ ID NO: 13 and SEQ ID NO: 17. The reverse primer has oligonucleotide sequences selected from the following groups: SEQ ID NO: 2, SEQ ID NO: 6, SEQ ID NO: 10, SEQ ID NO: 14 and SEQ ID NO: 18. The molecular beacon is configured to attach to a pathogen target sequence. The beacon has an oligonucleotide sequence selected from the group consisting of SEQ ID NO: 3, SEQ ID NO: 7, SEQ ID NO: 11, SEQ ID NO: 15, and SEQ ID NO: 19 and a fluorophore.
Owner:TELEFLEX MEDICAL INC
NASBA-microwell plate detection kit and detection method for five vibrios in mariculture
ActiveCN103834723BQuick checkEasy to operateMicrobiological testing/measurementAgainst vector-borne diseasesForward primerVibrio anguillarum
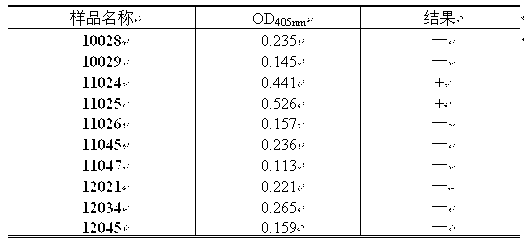
The invention relates to an NASBA-microwell plate detection kit and a detection method for five vibrios in mariculture. The kit comprises an enzyme mixed solution, a reaction liquid containing a forward primer and a reverse primer, and five probe solutions. The five probe solutions are a vibrio parahaemolyticus probe solution, a vibrio alginolyticus probe solution, a vibrio harveyi probe solution, a vibrio vulnificus probe solution and a vibrio anguillarum probe solution. The detection method comprises the steps of extracting total RNA by using a cracking liquid, amplifying the total RNA to obtain an NASBA amplification product by using the reaction solution; and at the same time, coating the probes in the microwell plate; subjecting the NASBA amplification product and the probes to hybrid chromogenic reaction; and then detecting absorption value of A405 nm. The detection kit is sensitive and specific, is convenient for operation in detection and can detect five vibrios in the mariculture rapidly.
Owner:NINGBO UNIV
Macrobrachium rosenbergii Nodavirus NASBA-LFD detection method and detection kit thereof
ActiveCN103409556BStrong specificityHigh detection sensitivityMicrobiological testing/measurementRNA extractionNucleic acid
The invention discloses a Macrobrachium rosenbergii Nodavirus NASBA-LFD (nuclear acid sequence-based amplification-lateral flow dipstick) detection method and a detection kit thereof. The detection method includes: according to a Macrobrachium rosenbergii Nodavirus (MrNV) sequence published by GenBank, screening a conserved region, and designing the primers and probe needed by an MrNV NASBA-LFD reaction system; employing a tissue RNA extraction reagent prepared by the inventor to extract the RNA of a to-be-detected sample, then utilizing the MrNV NASBA reaction system established by the invention to perform detection, and at the end of the reaction, determining the result according to nucleic acid rapid test paper. Compared with common detection technologies of the virus, the method and the kit provided in the invention have the characteristics of simplicity and rapidity, good specificity and high sensitivity, can be used for field service by technical personnel and farmers at the production front line, and are in favor of diagnosis prevention and pathogen blocking of Macrobrachium rosenbergii white tail disease.
Owner:ZHEJIANG INST OF FRESH WATER FISHERIES
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com