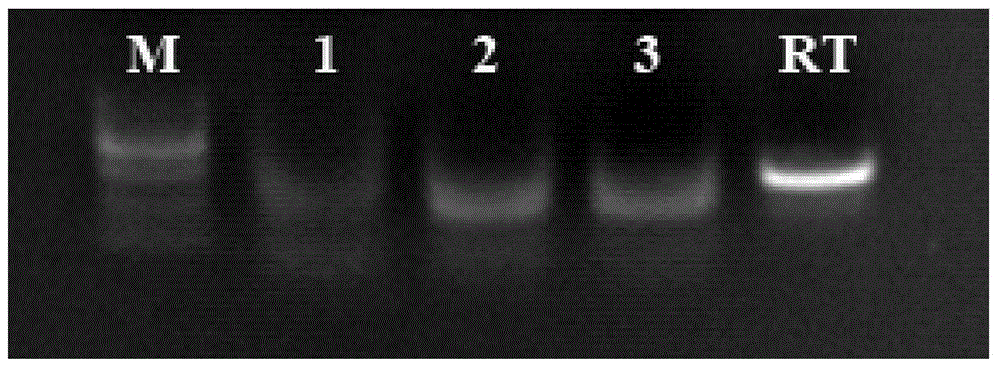
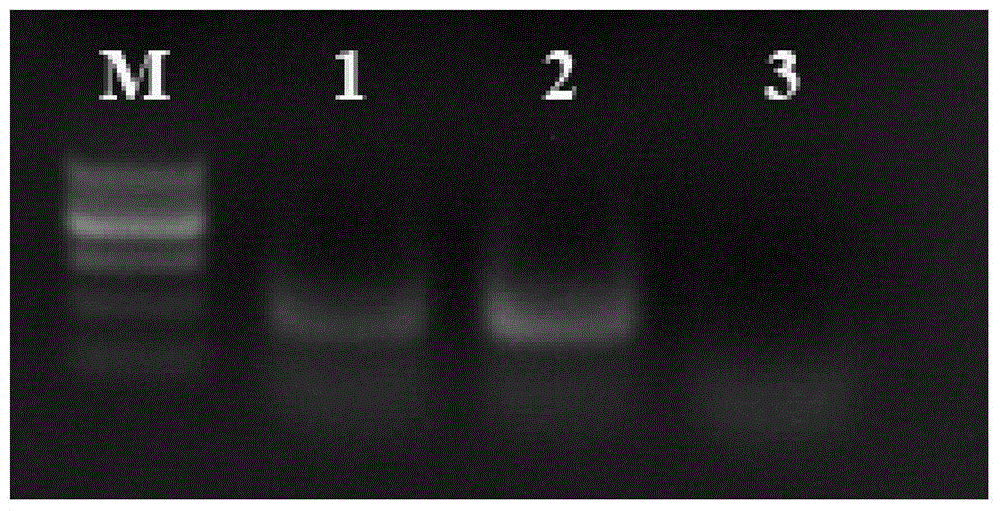
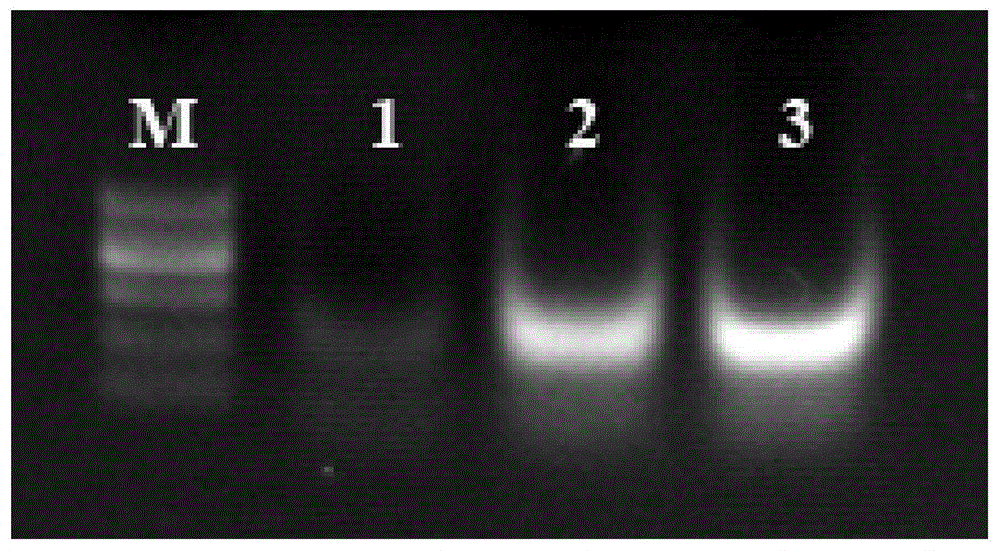
NASBA-ELISA (nucleic acid sequence-based amplification-enzyme-linked immuno sorbent assay) detection primer, probe and kit for type II grass carp reovirus
A technique for detecting reoviruses and primers, which is applied in the directions of DNA/RNA fragments, recombinant DNA technology, and microbial measurement/inspection. High detection efficiency and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1: Design and synthesis of specific primers and probes
[0039] The S6 sequences of seven strains of GCRV-GD108, 106, 918, HeNan988, HeNan794, HZ08 and 109 were obtained from NCBI and compared; a pair of amplification primers and corresponding capture probes and detection probes were designed in the conserved region.
[0040] S6-F: 5'- GATGCAAGGTCGCATATGAG CAGACGGGTCACCGTATTGTTACA-3' (SEQ ID NO.1), the underlined part is complementary to the detection probe;
[0041] S6-R: 5'- AATTCTAATACGACTCACTAAAGGGAGAAGG GAACTCATCAGTAAAGTCGGA-3' (SEQ ID NO.2), the underlined part is the T7 promoter sequence.
[0042] Type II GCRV S6 gene-specific capture probe: S6-CP: 5'BIO-TCAGCAGGTGCCGTTAATTTGT-3' (SEQ ID NO.3);
[0043] Type II GCRV S6 gene-specific detection probe S6-DP: 5'-TGATGCAAGGTCGCATATGAG-DIG3' (SEQ ID NO.4).
Embodiment 2
[0044] Exploration and Optimization of Primer Concentration, Enzyme Concentration and Ion Concentration in Example 2 NASBA Method
[0045] (1)K + Concentration optimization
[0046] In various literatures, K + Concentrations are 10mM, 20mM, 42mM, 70mM, 120mM; literature shows that the concentration above 100mM has little effect on the results; K in the basic system + The concentration is 25mM; the KCl concentration is respectively set to 25mM, 50mM, and 75mM for NASBA amplification. The results of 50mM and 75mM are similar, both of which are brighter than the band amplified by 25mM, so it is determined to use 50mM KCl in the future system.
[0047] (2) Optimization of the dosage of RNaseH
[0048] In different literatures, the amount of RNaseH used in each 20ul reaction system ranged from 0.1U, 0.2U to 0.8U; DMSO or sorbitol was added to the reaction system using less RNaseH to stimulate the endogenous RNaseH activity of AMV. However, too little RNaseH is difficult to abs...
Embodiment 3
[0053] The establishment of embodiment 3NASBA-ELISA reaction system and reaction program
[0054] 1. Extraction of viral RNA
[0055] ①Take 200ul virus sample, add 1ml Trizol, shake vigorously for 15s, and place at room temperature for 5min;
[0056] ② Add 200ul chloroform, shake vigorously for 15s, and centrifuge at 12000rpm at 4°C for 15min;
[0057] ③Take the supernatant, add 650ul isopropanol, and precipitate at -20°C for 10min;
[0058] ④ Centrifuge at 12000rpm at 4°C for 15min, remove the supernatant;
[0059] ⑤ Add 75% ethanol prepared with 1ml DEPC water to wash the precipitate, and centrifuge at 8000rpm at 4°C for 5min;
[0060] ⑥ Gently remove the supernatant, suck off the residual liquid with a pipette tip, and dry the precipitate at room temperature for 10 minutes;
[0061] ⑦ Add 24ul of DEPC water to dissolve the precipitate, divide 5ul into 8 tubes and store at -80°C for later use; at the same time, extract RNA from healthy grass carp tissue as a control.
...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com