Converting diploidy to haploidy for genetic diagnosis
a technology of genetic diagnosis and haploidy, applied in the field of haploidy conversion to haploidy for genetic diagnosis, can solve the problems of complicated interpretation of sequencing ladders, affecting the accuracy of sequencing results, so as to simplify the analysis of protein products
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
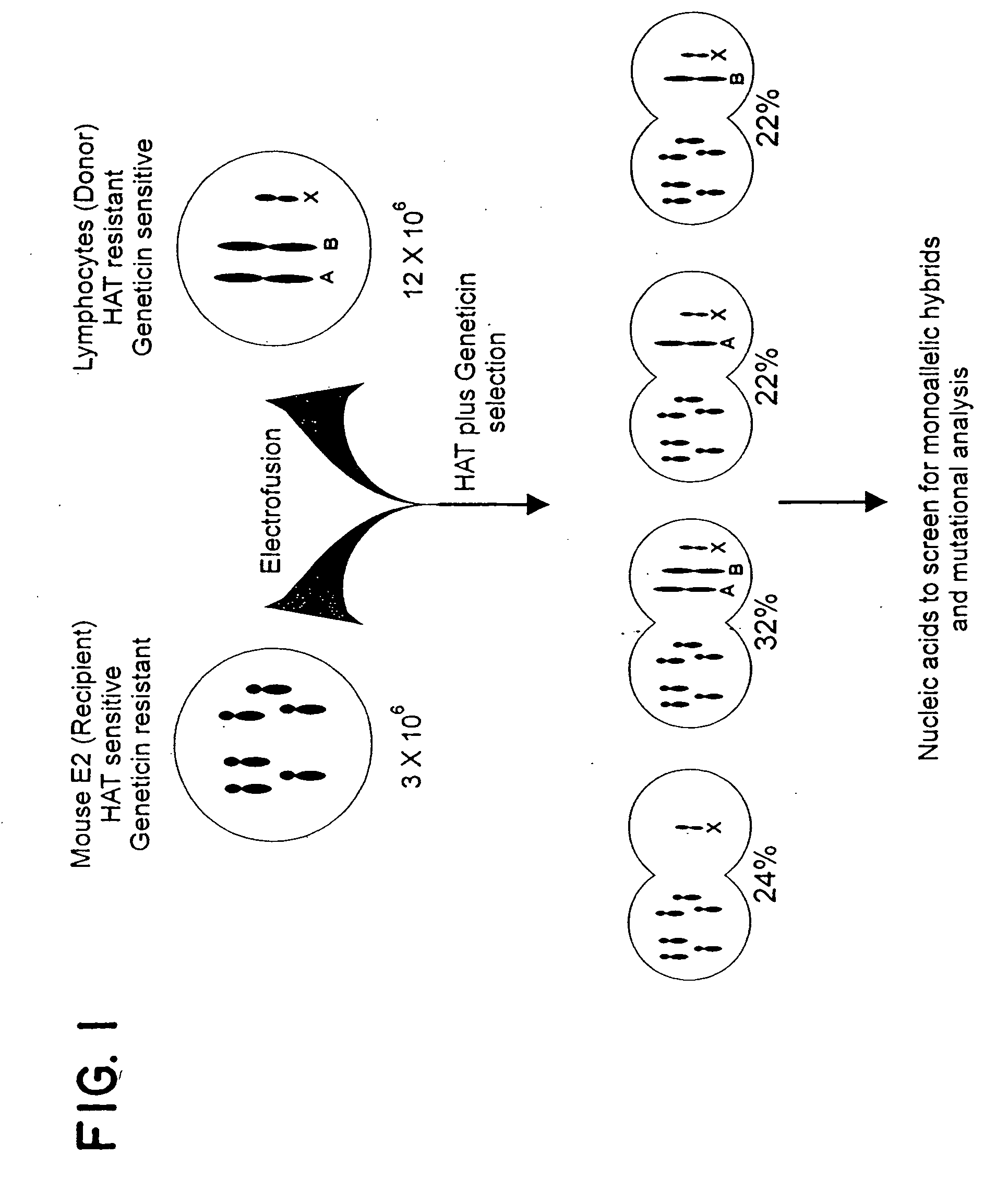
[0033] An outline of the approach is presented in FIG. 1. The rodent fusion partner was a line derived from mouse embryonic fibroblasts transformed with ras and adenovirus E1A oncogenes. HPRT-deficient subclones of this line were generated, and one subclone (E2) was chosen for further experimentation based on its robust growth characteristics, maintenance of diploidy, and fusion efficiency (10). Human lymphocytes cells were mixed with E2 cells at an optimum ratio and electrofused, and hybrids selected in geneticin (to kill unfused human cells) and HAT (to kill unfused E2 cells) (11). Colonies appearing after two weeks of growth were expanded and RNA and DNA prepared for analysis. From a single fusion experiment, an average of 36 hybrid clones were obtained (range of 17 to 80 in five different individuals).
[0034] All hybrids contained the human X chromosome, as this chromosome contains the HPRT gene allowing growth in HAT. To determine whether other human chromosomes were present in...
example 2
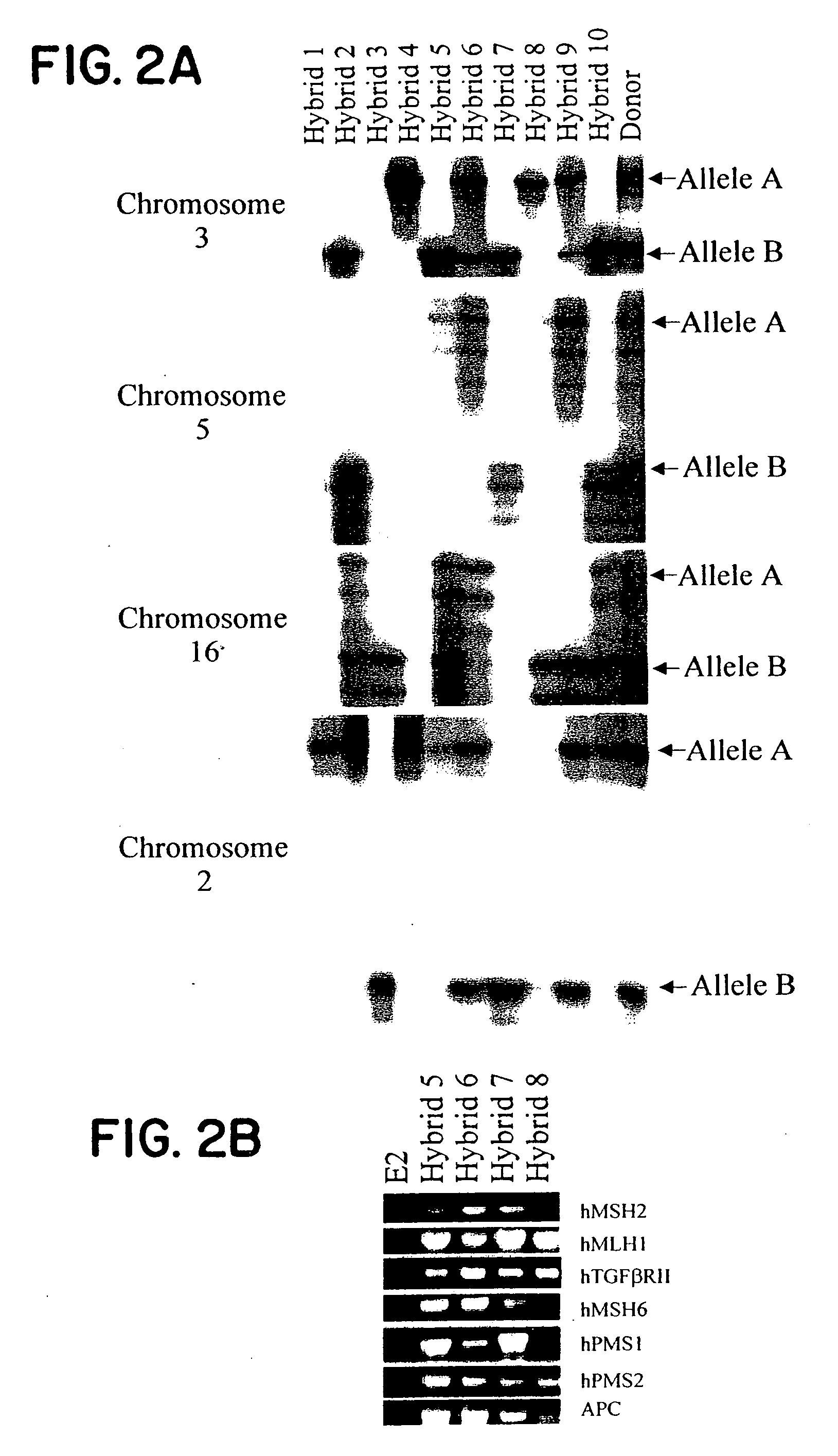
[0035] Two other features of the hybrids were essential for the analyses described below. First, the human chromosome complements of the hybrids were remarkably stable. Polymorphic marker analysis in ten hybrids revealed identical patterns of retention after growth for 90 (30 passages) generations after initial genotyping. Second, those hybrids containing the relevant chromosome expressed every human gene assessed, including all known colorectal cancer susceptibility genes (the hMSH2 and hMSH6 genes on chromosome 2p, the hPMS1 gene on chromosome 2q, the TGF-β Receptor Type II gene and hMLH1 gene on chromosome 3p, the APC gene on chromosome 5q, the hPMS2 gene on chromosome 7q, and the E-cadherin gene on chromosome 16q; representative examples in FIG. 2B) (14).
example 3
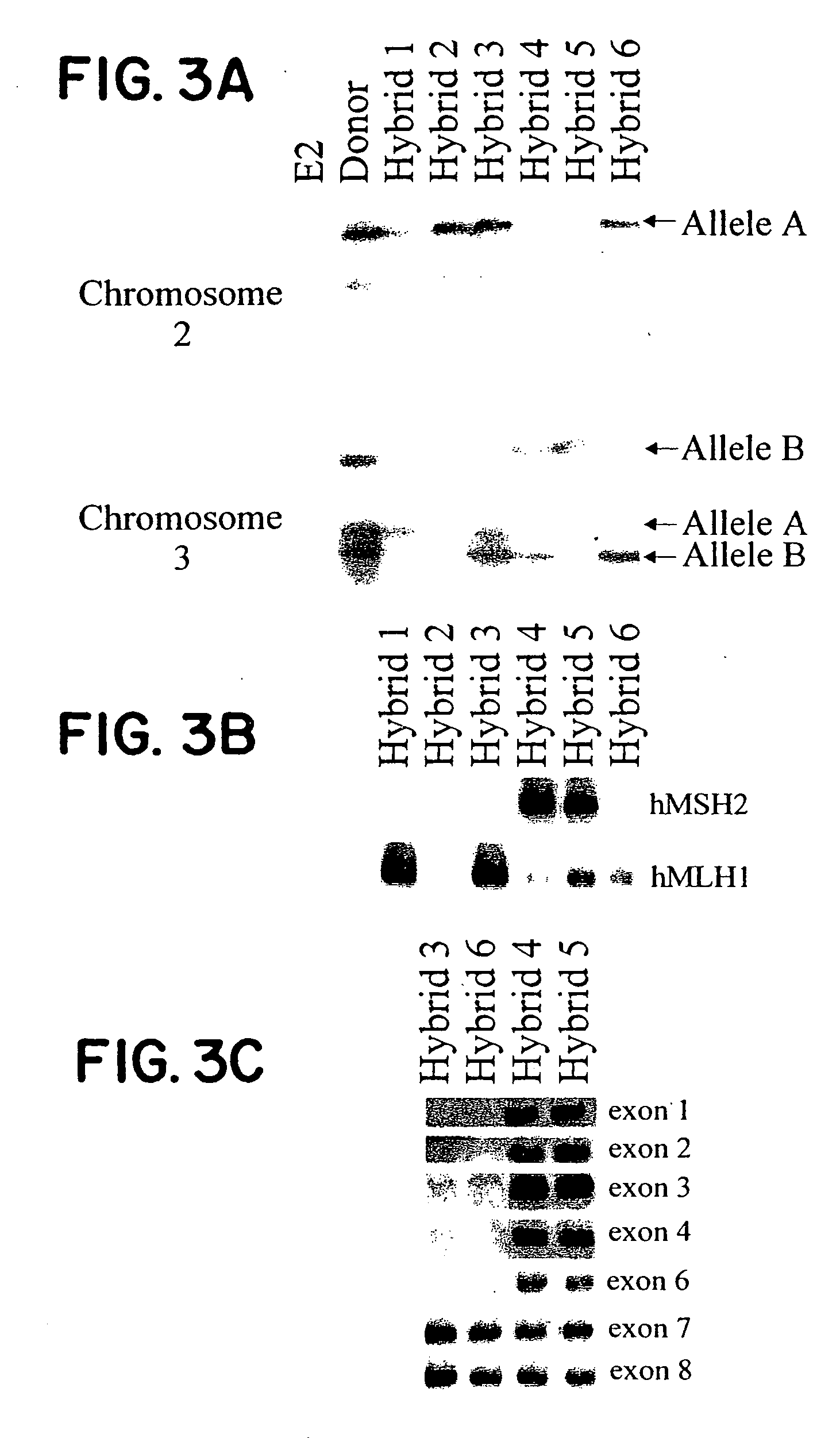
[0036] Having established the stability and expression patterns of CRC-predisposition genes in these hybrids, we used this “conversion” approach to investigate ten patients who had proven refractory to standard genetic diagnostic techniques. Each of these patients had a significant family history of colorectal cancer and evidence of mismatch repair deficiency in their tumors, yet sequencing of the entire coding sequence of each known MMR gene had failed to reveal mutations. Indeed, these and similar studies have prompted the speculation that other major HNPCC genes must exist. (25-34) Hybrids were generated from lymphocytes of each patient, and at least one hybrid containing the maternal allele and one hybrid containing the paternal allele of each MMR gene was isolated. Analysis of the nucleic acids from these hybrids revealed specific mutations in all ten patients (Table 1). In every case, an abnormality was found in a single allele of either hMSH2 or hMLH1. The nature of the abnor...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| polymorphic | aaaaa | aaaaa |
| disorder | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com