Liver tumor marker sequences
a marker sequence and liver tumor technology, applied in the field of liver tumor marker sequences, can solve the problems of lack of successful treatment options and no mutational model developed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example
Materials and Methods
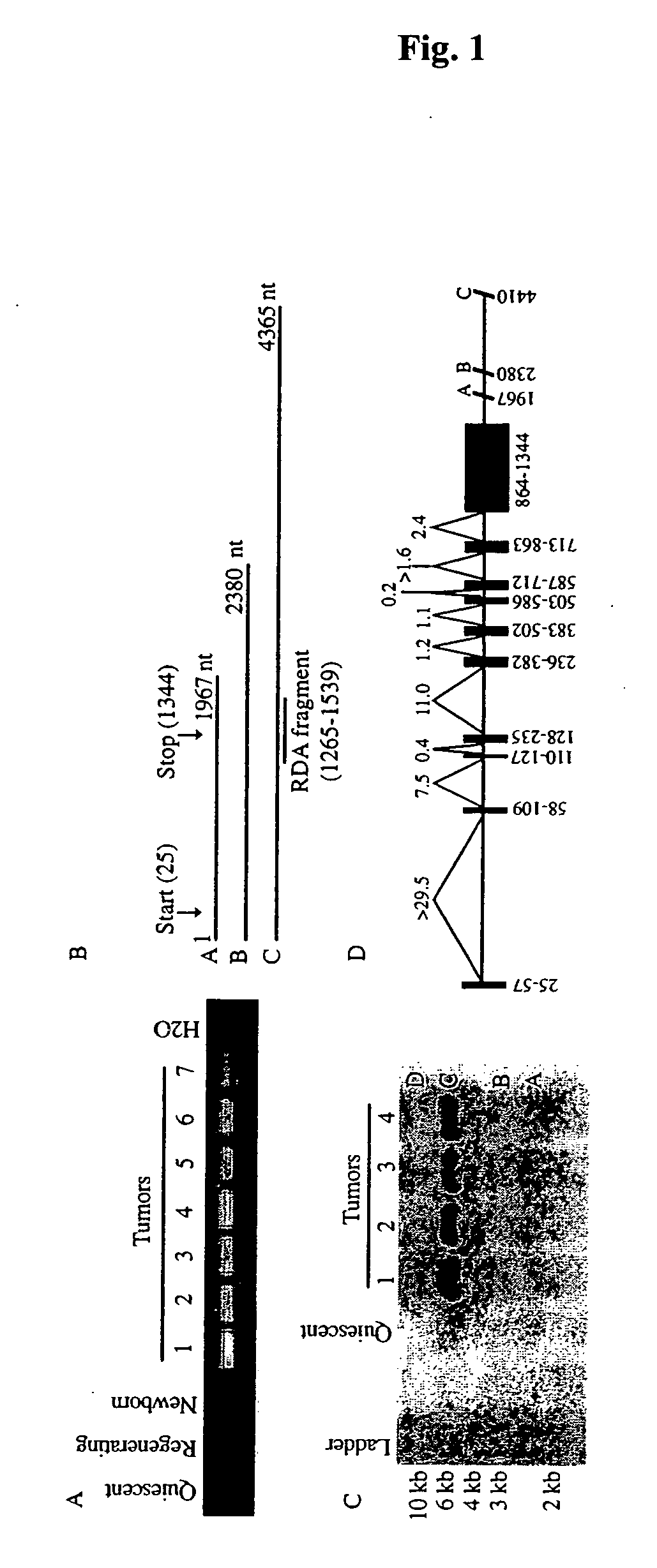
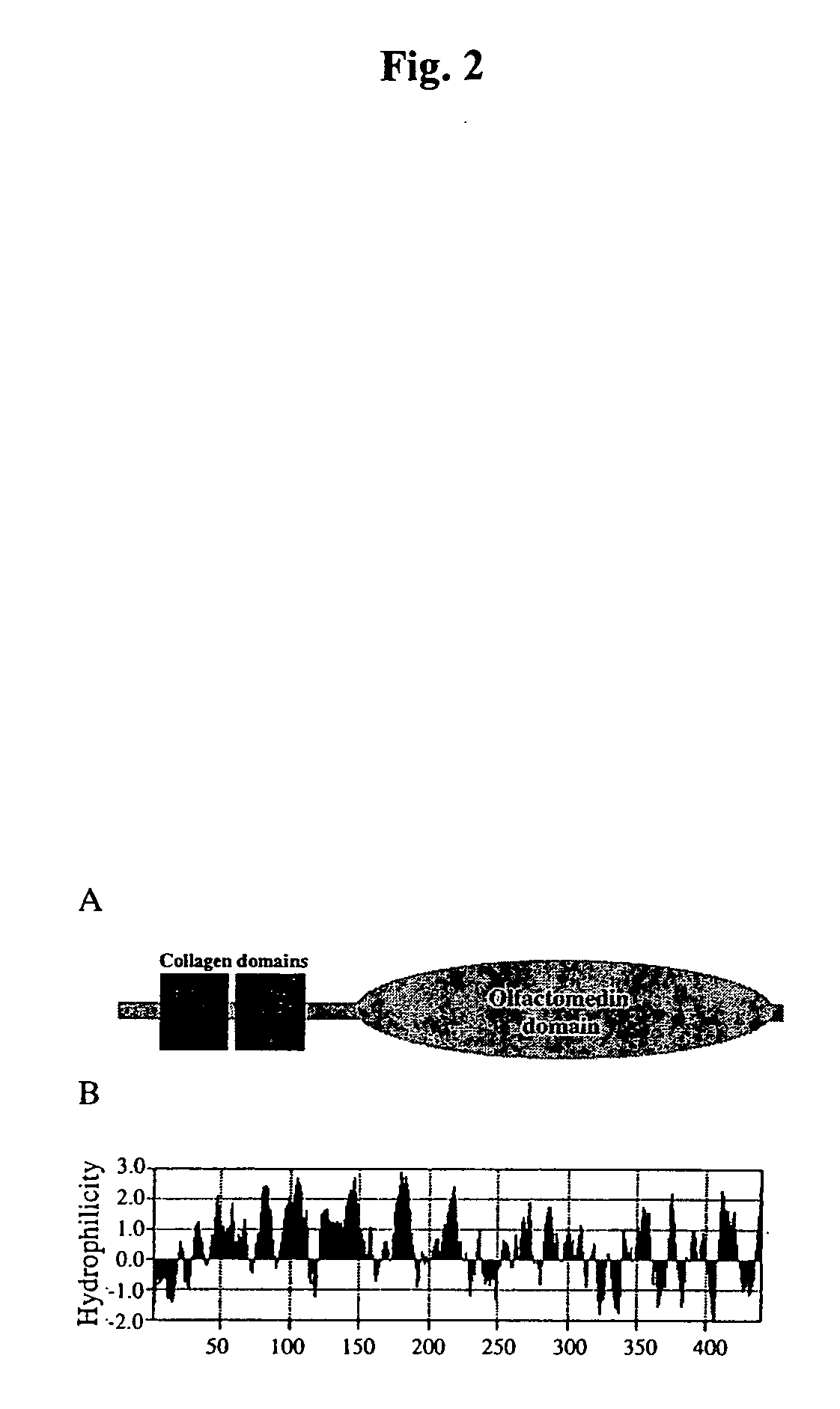
[0039] Rapid Amplification of cDNA Ends (RACE). Rapid amplification of cDNA ends (RACE) was performed in both directions using the SMART cDNA amplification kit (Clontech) from mouse liver tumor polyA RNA. 5′ and 3′ RACE were performed using the gene-specific primers, GSP-A [5′-GCATGGCAAGAACAGACTGG-3′] (SEQ ID NO:5) and GSP-B [5′-GGATGAGAAGGGCATCTGGA-3′] (SEQ ID NO:6). 5′ and 3′ RACE products that were identified with the corresponding GSP primer were gel extracted and cloned into TOPO-TA vector (Invitrogen). Cloned products were sequenced by Big Dye (ABI) in the McArdle Laboratory Sequencing Facility (University of Wisconsin-Madison).
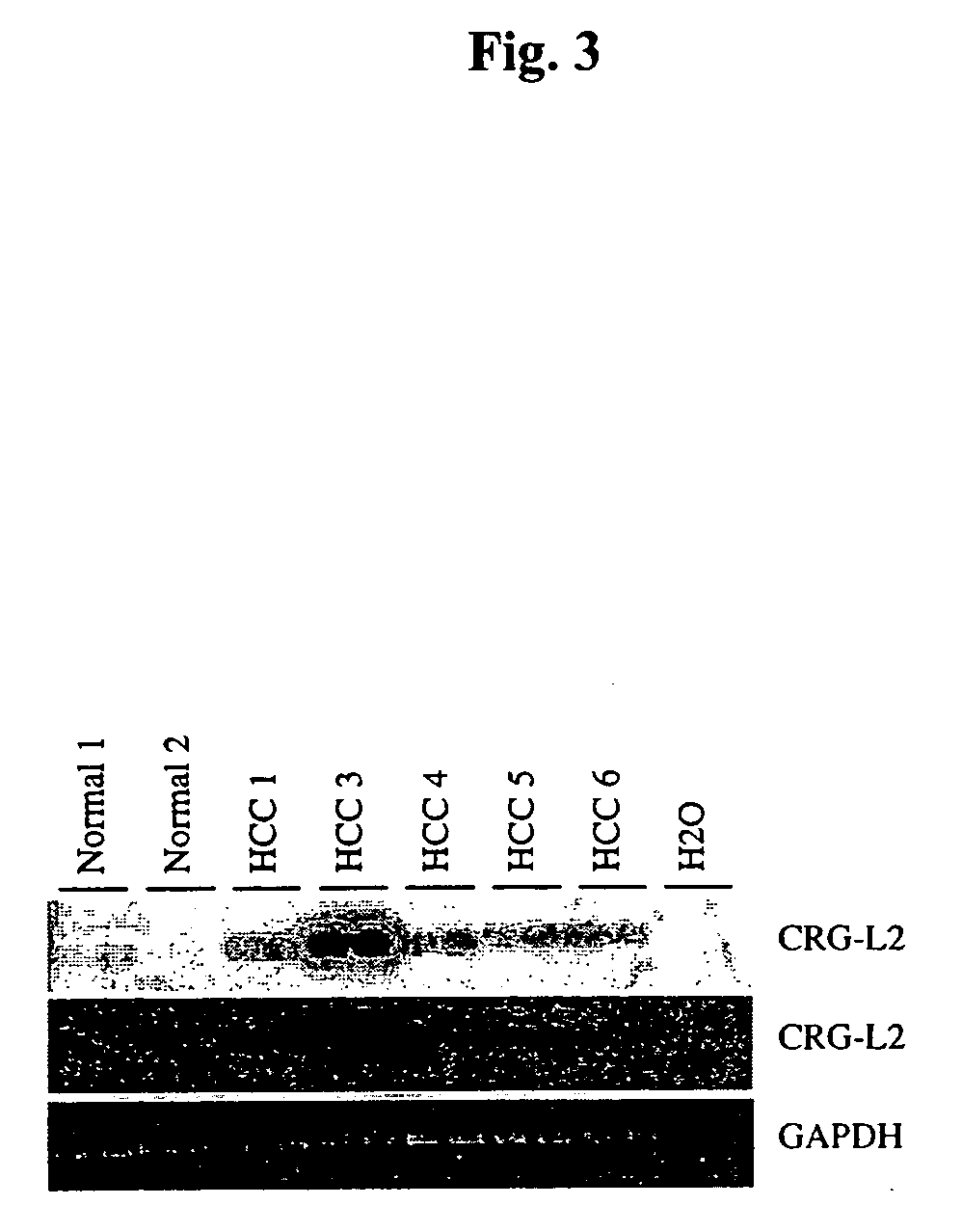
[0040] RNA Analysis. For analysis of murine CRG-L2 mRNA, total RNA was extracted from liver using guanidine thiocyanate / CsCl as described previously in (Lukas et al., 1999, incorporated by reference in its entirety). PolyA mRNA was isolated from 250 μg of total RNA using Oligotex mRNA Kit (Qiagen). RT-PCR was performed as describ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| mRNA structure | aaaaa | aaaaa |
| Northern blot | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com