CD52 OPTIMIZED Fc VARIANTS AND METHODS FOR THEIR GENERATION
a variant and variant technology, applied in the field of optimized variants, can solve the problems of unsatisfactory potency of antibodies as anti-cancer agents, another level of complexity, and the optimization of antibodies for clinical use, and achieve the effect of greater potential
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Computational Screening and Design of Fc Libraries
[0191] Computational screening calculations were carried out to design optimized Fc variants. Fc variants were computationally screened, constructed, and experimentally investigated over several computation / experimentation cycles. For each successive cycle, experimental data provided feedback into the next set of computational screening calculations and library design. All computational screening calculations and library design are presented in Example 1. For each set of calculations, a table is provided that presents the results and provides relevant information and parameters.
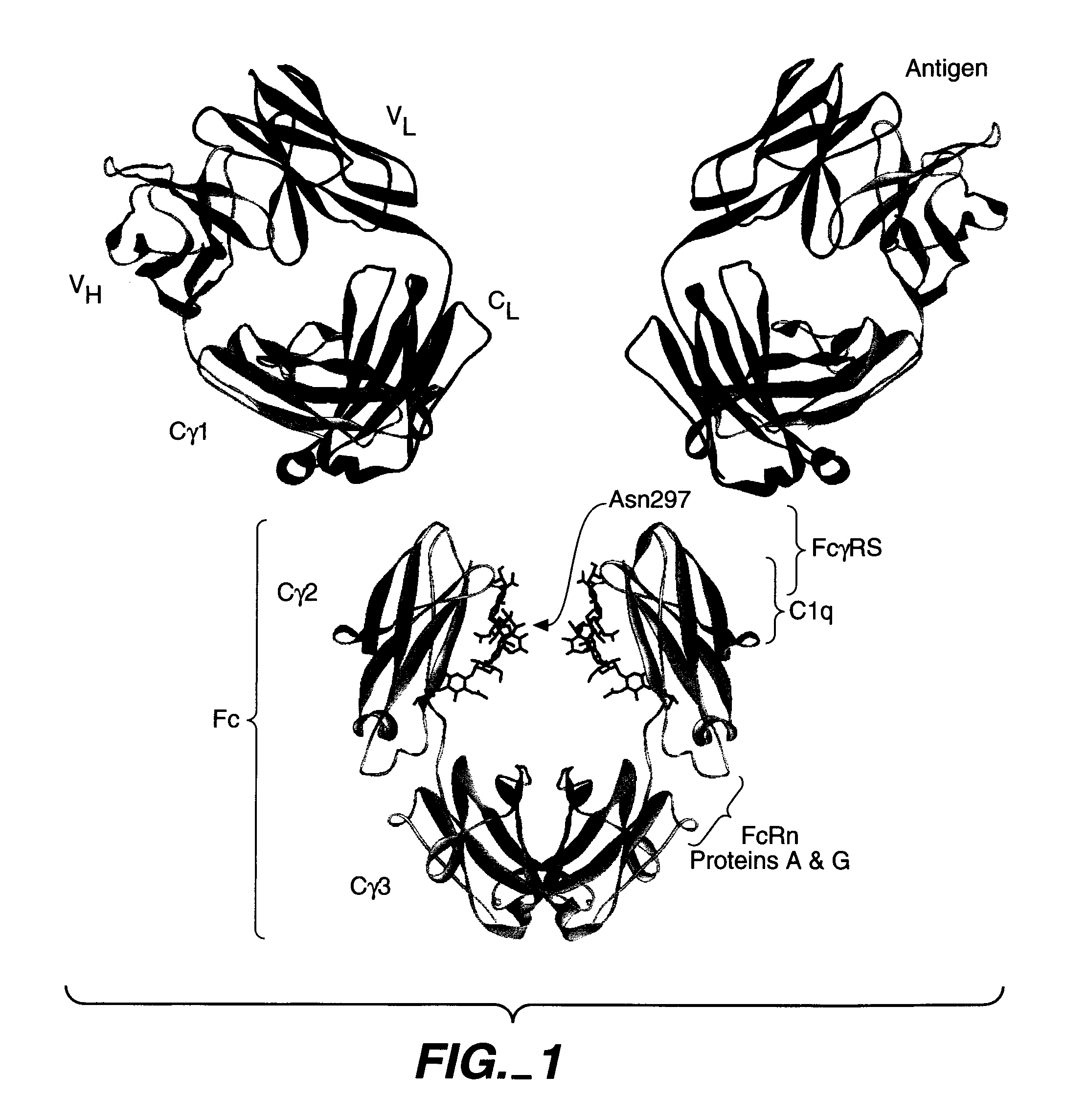
[0192] Several different structures of Fc bound bound to the extracellular domain of FcγRs served as template structures for the computational screening calculations. Publicly available Fc / FcγR complex structures included pdb accession code 1E4K (Sondermann et al., 2000, Nature 406:267-273.), and pdb accession codes 1IIS and 1IIX (Radaev et al., 2001, J Biol...
example 2
Experimental Production and Screening of Fc Libraries
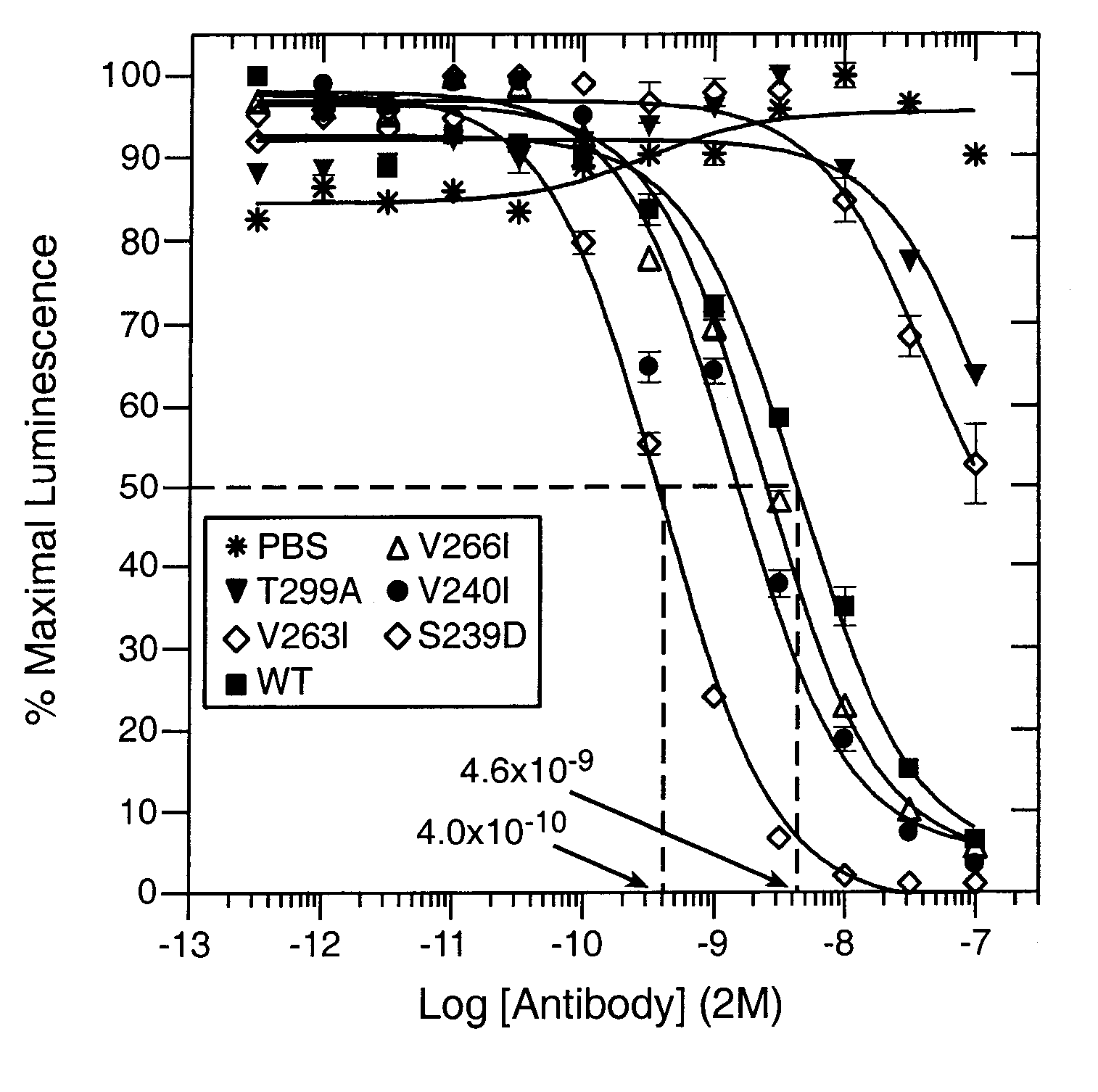
[0258] The majority of experimentation on the Fc variants was carried out in the context of the anti-cancer antibody alemtuzumab (Campath®, a registered trademark of Ilex Pharmaceuticals LP). Alemtuzumab binds a short linear epitope within its target antigen CD52 (Hale et al., 1990, Tissue Antigens 35:118-127; Hale, 1995, Immunotechnology 1:175-187). Alemtuzumab has been chosen as the primary engineering template because its efficacy is due in part to its ability to recruit effector cells (Dyer et al., 1989, Blood 73:1431-1439; Friend et al., 1991, Transplant Proc 23:2253-2254; Hale et al., 1998, Blood 92:4581-4590; Glennie et al., 2000, Immunol Today 21:403-410), and because production and use of its antigen in binding assays are relatively straightforward. In order to evaluate the optimized Fc variants of the present invention in the context of other antibodies, select Fc variants were evaluated in the anti-CD20 antibody rituxi...
example 3
Selectively Enhanced Binding to FcγRs
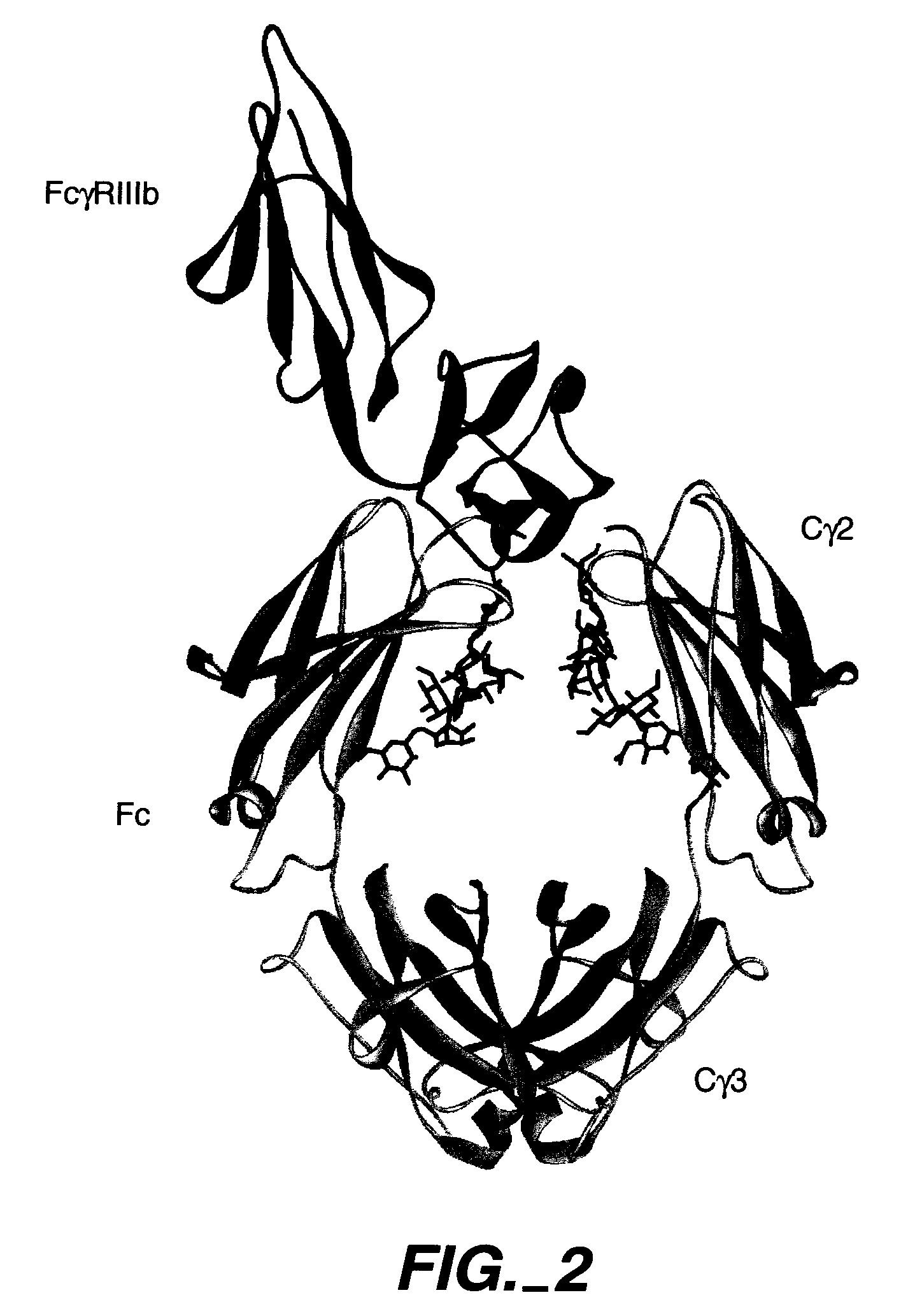
[0264] A number of promising Fc variants with optimized properties were obtained from the FcγRIIIa and FcγRIIb screen. Table 62 provides Fc variants that bind more tightly to FcγRIIa, and thus are candidates for improving the effector function of antibodies and Fc fusions. These include a number of variants that comprise substitutions at 239, 264, 272, 274, 330, and 332. FIGS. 13a and 13b show AlphaScreen™ binding data for some of these Fc variants. The majority of these Fc variants provide substantially greater FcγRIIIa binding enhancements over S298A / E333A / K334A (SEQ ID NO:45).
TABLE 62FcγIIIa-fold;FcγRIIIaFcγRIIbFcγIIb-VariantSEQ ID NOSubstitution(s)FoldFoldfold 1SEQ ID NO:9V264A0.53 2SEQ ID NO:10V264L0.56 3SEQ ID NO:11V264I1.43 4SEQ ID NO:12F241W0.29 5SEQ ID NO:13F241L0.26 6SEQ ID NO:14F243W0.51 7SEQ ID NO:15F243L0.51 8SEQ ID NO:16F241L / F243L / V262I / V264I0.09 9SEQ ID NO:17F241W / F243W0.07 10SEQ ID NO:18F241W / F243W / V262A / V264A0.04 11S...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| Energy Refinement | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com