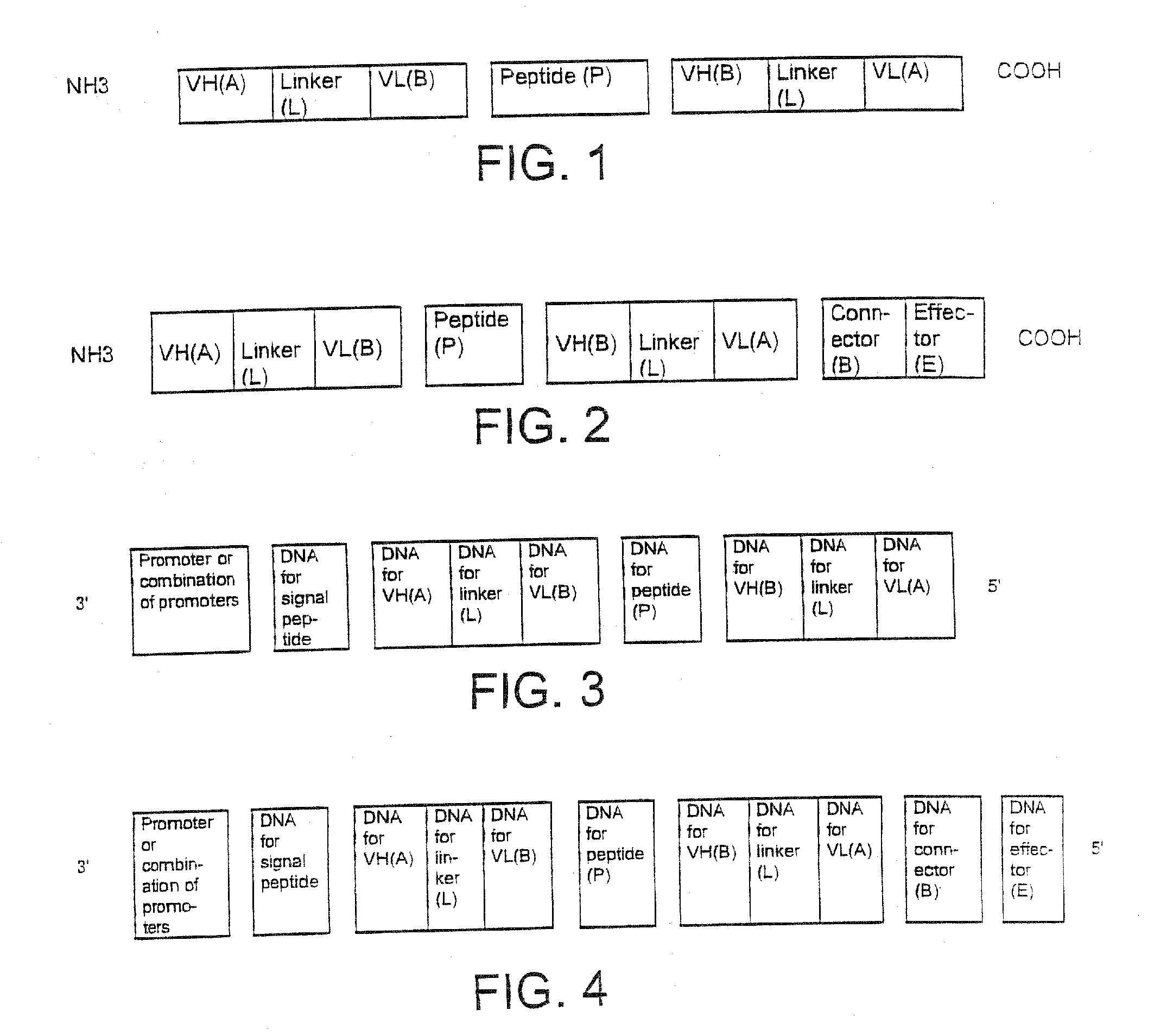
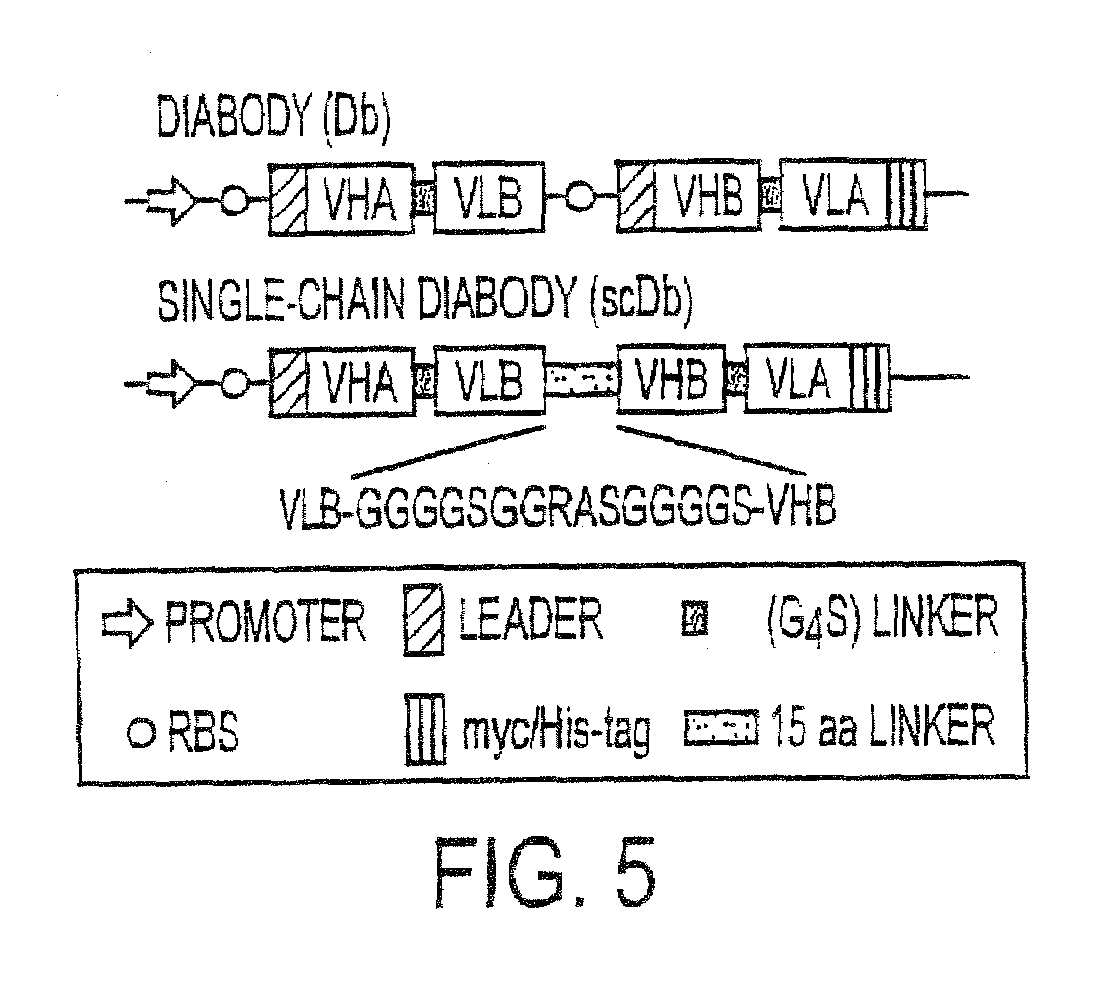
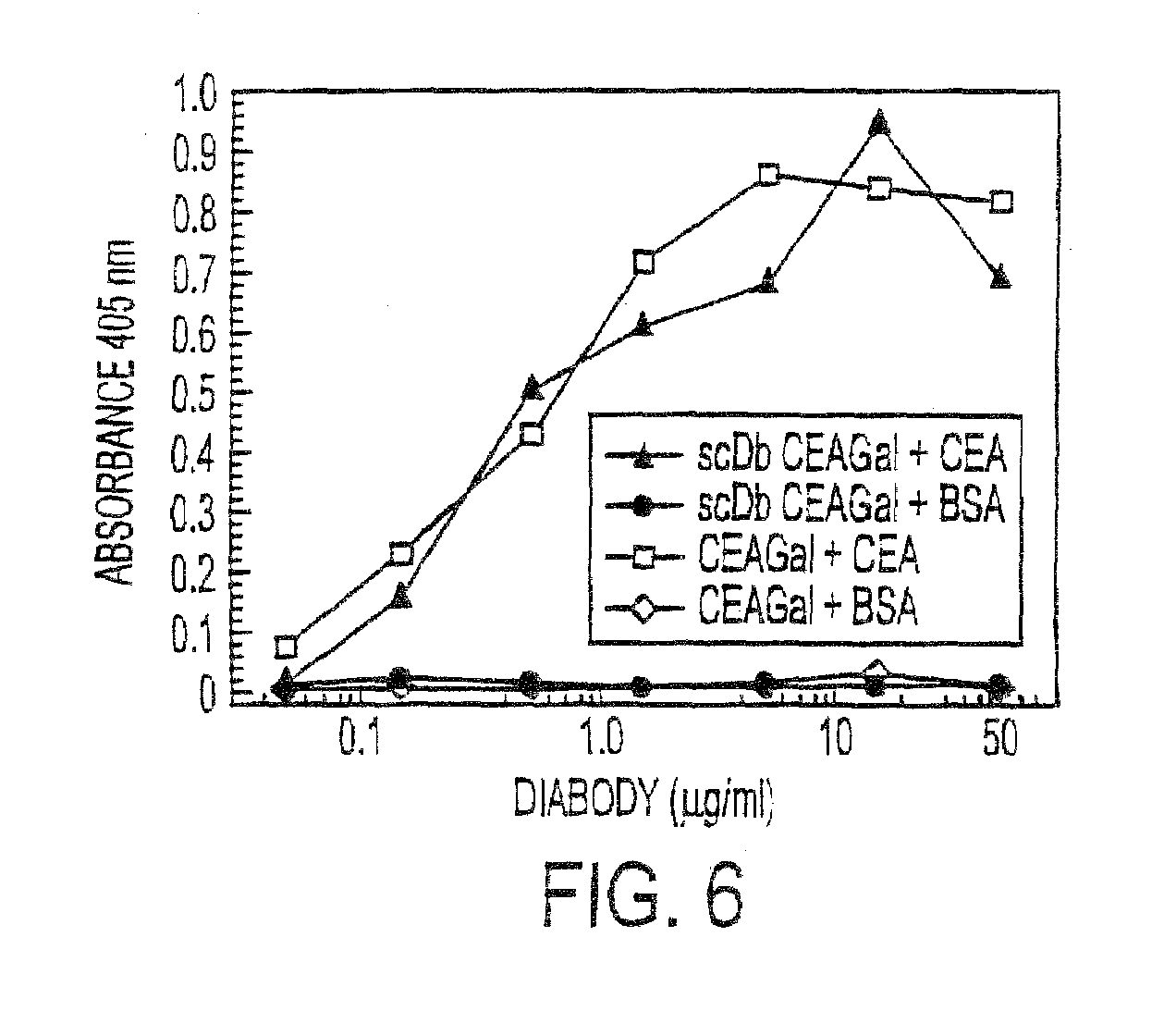
Single-chain Multiple Antigen-binding Molecule, Its Preparation and Use
a single-chain, antigen-binding technology, applied in the field of single-chain multiple antigen-binding molecules, can solve the problems of non-functional aggregates, non-functional associations of the four variable domains, and the selection of desired heterodimer molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation and Bacterial Expression of a Single-Chain Double Antigen-Binding Protein
[0169]The preparation of a single-chain double antigen-binding protein is described using the example of a protein which recognizes the antigens carcinoembryonic antigen (CEA) and E. coli β-galactosidase.
[0170]The following DNA sequences were connected together in the 5′ to 3′ direction as follows:[0171]LacZ promoter[0172]bacterial ribosome binding structure (AAGGAG)[0173]bacterial signal sequence pelB (Power et al., Gene 113, 95-99 (1992))[0174]VH anti-CEA (Kontermann et al., Immunotechnol. 3, 137 (1997))[0175]linker GGGS (SEQ ID NO.:19) (Kontermann et al., (1997))[0176]VL anti-β-galactosidase (Kontermann et al., (1997))[0177]connecting peptide GGGGSGGRASGGGGS (SEQ ID NO:3)[0178]VH anti-β-galactosidase (Kontermann et al., (1997))[0179]linker GGGGS (SEQ ID NO:1)[0180]VL anti-CEA (Kontermann et al., (1997))[0181]Myc epitope for antibody 9E10 EQKLISEEDLN (SEQ ID NO:8) (Munro & Pelham, Cell 46, 291-300...
example 2
Eukaryotic Expression of a Single-Chain Double Antigen-Binding Protein
[0190]For expression in eukaryotic cells, the coding region of the single-chain double antigen-binding protein was cloned into a eukaryotic expression vector (pSecTagA, Invitrogen), the bacterial signal sequence having been replaced by the Ig-κ signal sequence already present in the vector.
[0191]For this, the single-chain construct was amplified using the primers LMB2 and pelB-Metminus, whereby methionine in the pelB leader (position 21) was replaced by threonine. This was followed by hydrolysis with SfiI and EcoRI and cloning into the vector pSecTagA. The mature single-chain antigen-binding protein contains 7 additional amino acids (AAQPATA) (SEQ ID NO:14) at the N terminus of the VHA domain.
[0192]The plasmid was transiently transfected with Lipofectamine (Gibco) into eukaryotic HEK 293 cells. Stable cells were selected in the presence of zeocin. It was possible in immunofluorescence experiments with these cells ...
example 3
In Vitro Enzyme Recruitment by Cocultivation with Cells Secreting Single-Chain Double Antigen-Binding Protein
[0193]Recruitment was investigated in vitro by cocultivation of HEK 293 cells producing the single-chain double antigen-binding protein and CEA-positive LoVo cells. For this, firstly the cells producing the protein according to the invention were cultivated with the LoVo cells in Transwell cell culture dishes (Costar) in which the two cell lines are separated by a membrane. After two days, β-galactosidase (10 μg / ml) was added, and the recruitment was detected by adding the substrate X-Gal. A specific staining was found on cocultivation with the cells producing the single-chain double antigen-binding protein, whereas control experiments with untransfected HEK 293 cells showed no staining.
[0194]Experiments in which the LoVo cells were replaced by A549 cells (CEA-negative), and experiments in which the cells producing the single-chain double antigen-binding protein were incubate...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| nucleic acid | aaaaa | aaaaa |
| structure | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap