Soluble CTL A4 mutant molecules and use thereof
A mutant and molecular technology, applied in the field of soluble CTLA4 molecules, which can solve problems such as affinity effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
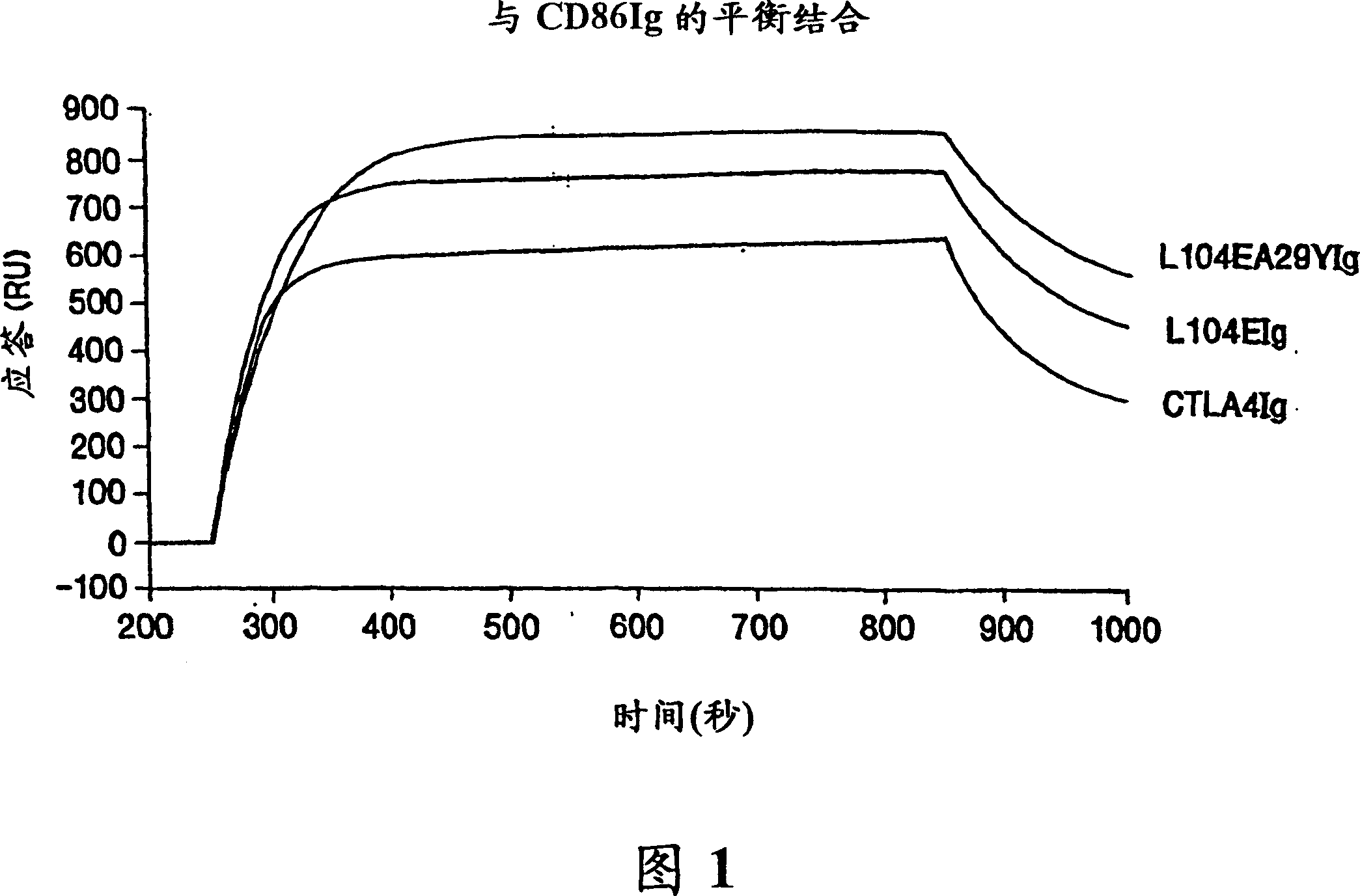
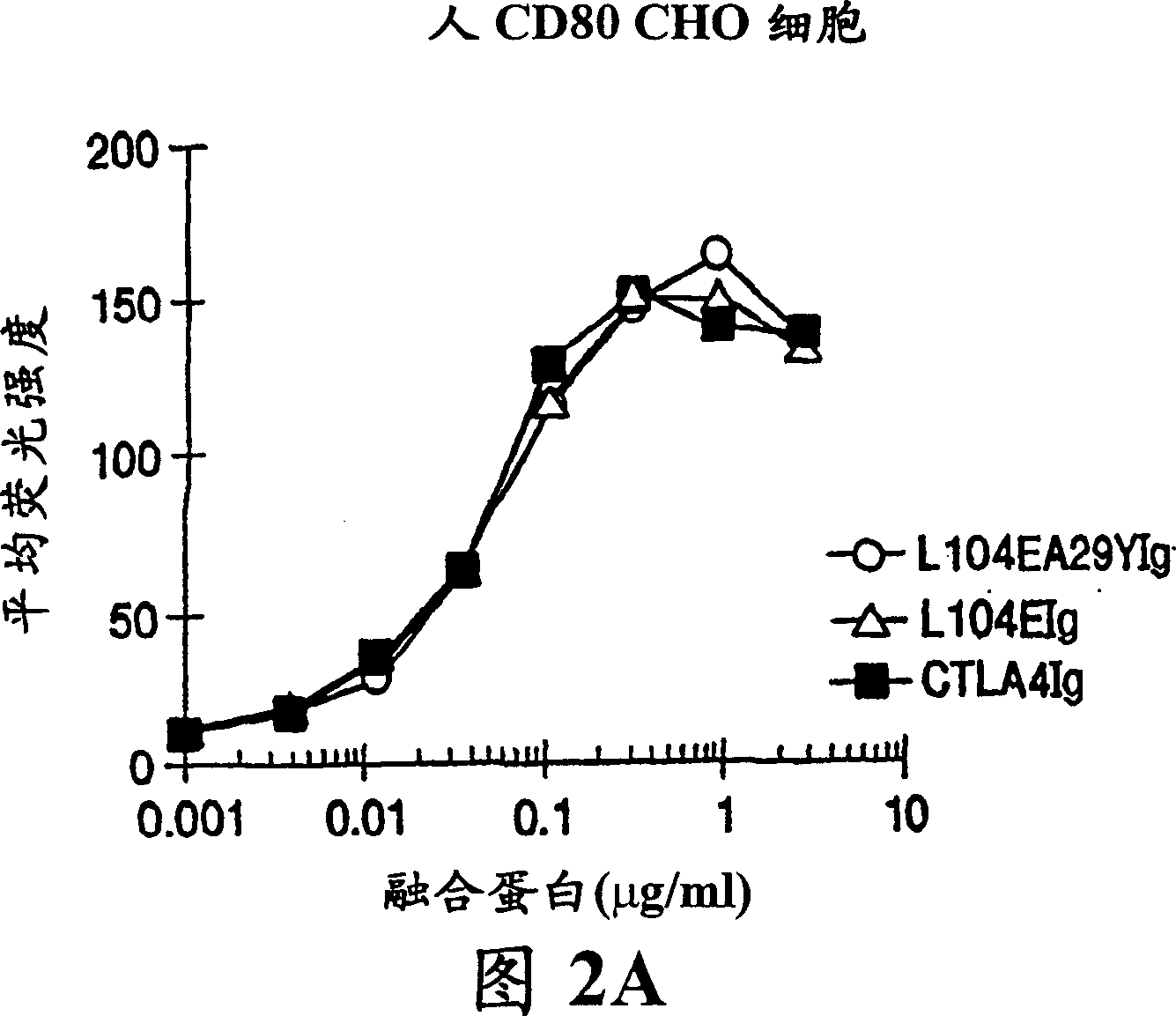
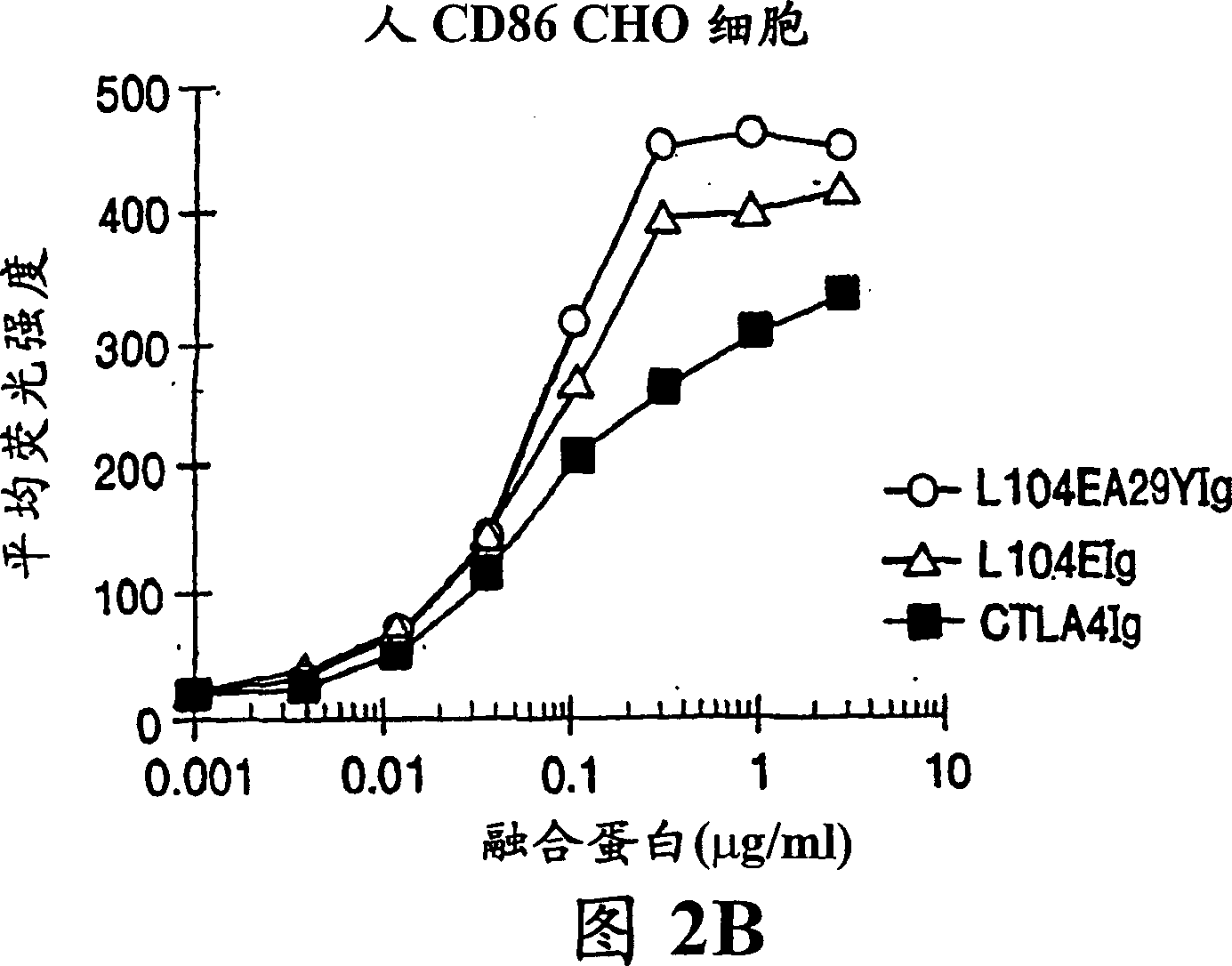
[0115] This example provides an illustration of the method used to generate a nucleotide sequence encoding a soluble CTLA4 mutant molecule of the invention. Single point mutant L104EIg was generated and tested for binding kinetics to CD80 and / or CD86. The L104EIg nucleotide sequence was used as a template to generate the double-site mutant CTLA4 sequence-L104EA29YIg, and the binding kinetics of L104EA29YIg to CD80 and / or CD86 was tested.
[0116] CTLA4Ig codon-based mutagenesis:
[0117] A mutagenesis and screening strategy was developed to identify mutant CTLA4Ig molecules with slower dissociation rates ("off" rates) from CD80 and / or CD86 molecules. Single point mutant nucleotide sequences were generated using CTLA4Ig (US Patent Nos. 5,844,095, 5,851,795, and 5,885,796; ATCC Deposit No. 68629) as a template. Designed for random mutagenesis of specific cDNA codons by allowing any base in codon positions 1 and 2, but only guanine and thymine in position 3 (XXG / T; also known a...
Embodiment 2
[0120]A description of the screening method used to identify the single-site and double-site mutant CTLA4 polypeptides expressed from the constructs described in Example 1, wherein the mutant CTLA4 polypeptides exhibit binding affinity for the CD80 and CD86 antigens, is provided below Higher than non-mutated CTLA4Ig molecules.
[0121] Current in vitro and in vivo studies demonstrate that CTLA4Ig by itself cannot completely block the priming of antigen-specific activated T cells. In vitro studies measuring inhibition of T cell proliferation using CTLA4Ig and CD80 or CD86 specific monoclonal antibodies showed that anti-CD80 monoclonal antibodies did not enhance CTLA4Ig inhibition. Anti-CD86 monoclonal antibodies, however, enhanced the inhibition, suggesting that CTLA4Ig was not as effective in blocking CD86 interactions. These data support the earlier findings of Linsley et al. ( Immunity , (1994), 1:793-801), the finding that inhibition of CD80-mediated cellular responses r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com