SAB as a Biomarker for Degenerative Diseases and Therapeutic Sensitivity in Cancers
a biomarker and cancer technology, applied in the field of blood factors, can solve the problems attracting death-inducing proteins to the mitochondrial surface, etc., and achieve the effects of reducing metabolic efficiency, monitoring and modulating mitochondrial function, and destabilizing integrity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
In-Cell Western Blot Analysis
[0148]Cells or tissue samples are placed in a fluorescent compatible plate (for example, a 96-well plate) and the sample can be fixed to the plate. The sample is then permeabilized using detergent to disrupt the plasma membrane. Primary antibodies to detect SAB (Abnova, Inc.) are then incubated with the samples. Following a brief wash, the cells are incubated with a fluorescently labeled secondary antibody used to recognize the primary antibody. The samples are washed again. The plate is then scanned on a Li-Cor Bioscience Odyssey CLx near-infrared scanner to measure the fluorescent signal from the sample. A high fluorescent signaling indicates a higher level of SAB protein, while a low fluorescence signal indicates a lower level of SAB protein.
[0149]This assay can be used to detect SAB protein concentrations in a variety of cells and tissues. The assay can be useful for adherent cells, non-adherent cell, and primary cells. The assay can be modified to d...
example 2
SAB Levels in Various Disease Conditions
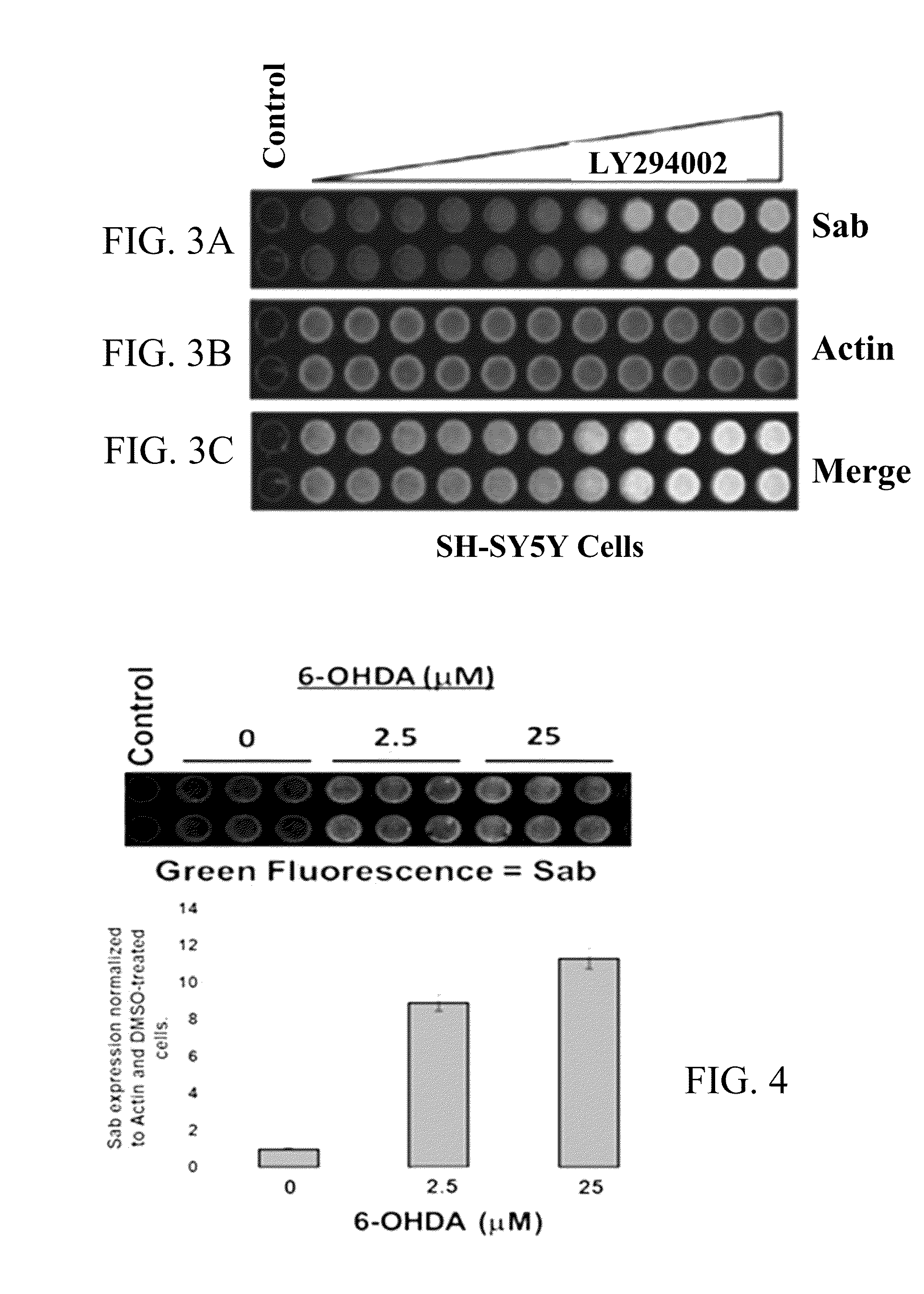
[0150]Parkinson's disease-like state was induced in animals by localizing treatment to the ventral midbrains of rats using 6-hydroxydopamine. Blood was obtained from 6-hydroxydopamine treated rats and the control rats. Levels of SAB protein or RNA encoding SAB proteins in platelets and other blood cells were higher in 6-hydroxydopamine treated rats compared to control rats.
[0151]Similarly, blood samples can be obtained from humans suffering from Parkinson's disease and humans free from Parkinson's disease. Levels of SAB protein or RNA encoding SAB proteins in platelets and other blood cells will be higher in the humans suffering from Parkinson's disease compared to the humans free from Parkinson's disease.
[0152]Tissues samples from ventral midbrain and muscles were obtained from young rats and aged rats. SAB protein levels were determined in the two sets of samples. SAB levels were higher in the ventral midbrains and muscles of aged rats compa...
example 3
Correlation Between SAB Expression, Mitochondrial Priming, and Susceptibility of a Cancer to Apoptosis Inducing Chemotherapeutic Agents
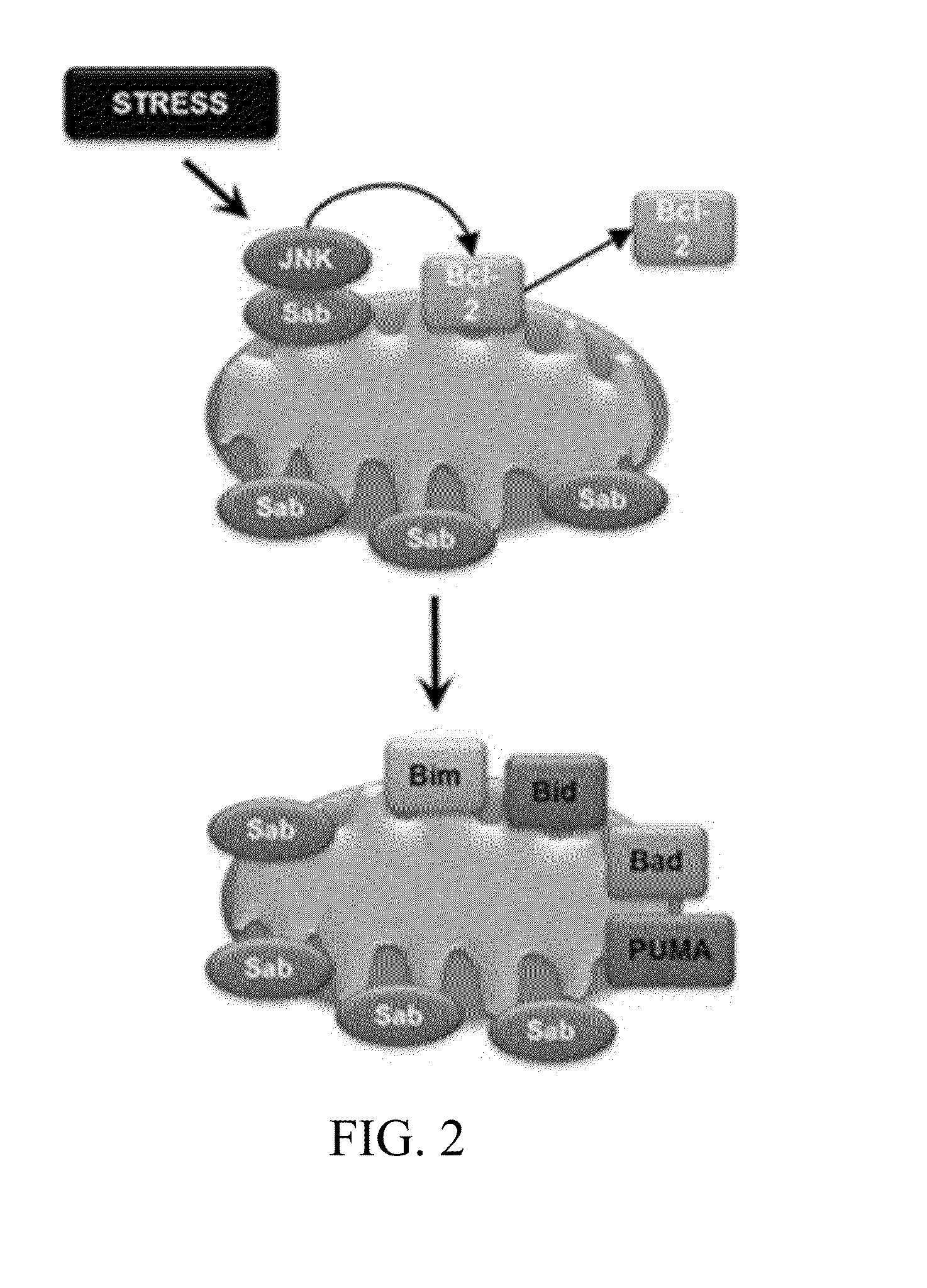
[0155]Mitochondrial priming influences cancer cell responses to chemotherapy. Mitochondrial priming is the process potentiating mitochondrial physiology towards a pro-apoptotic response. In cancer cells, mitochondrial priming has been used as means to sensitize cells to chemotherapy and radiotherapy approaches. Mitochondrial priming drives cancer cells toward apoptosis by exploiting their unique biology and permits physicians to administer lower doses of toxic chemotherapies. The amount of mitochondrial priming can be used as a predictive index of chemotherapeutic success. Elevated levels of pro-apoptotic Bcl-2 family members are seen on primed mitochondria. Therefore, detection of primed mitochondria in tumors can be used as a biomarker for chemotherapeutic success (FIG. 10).
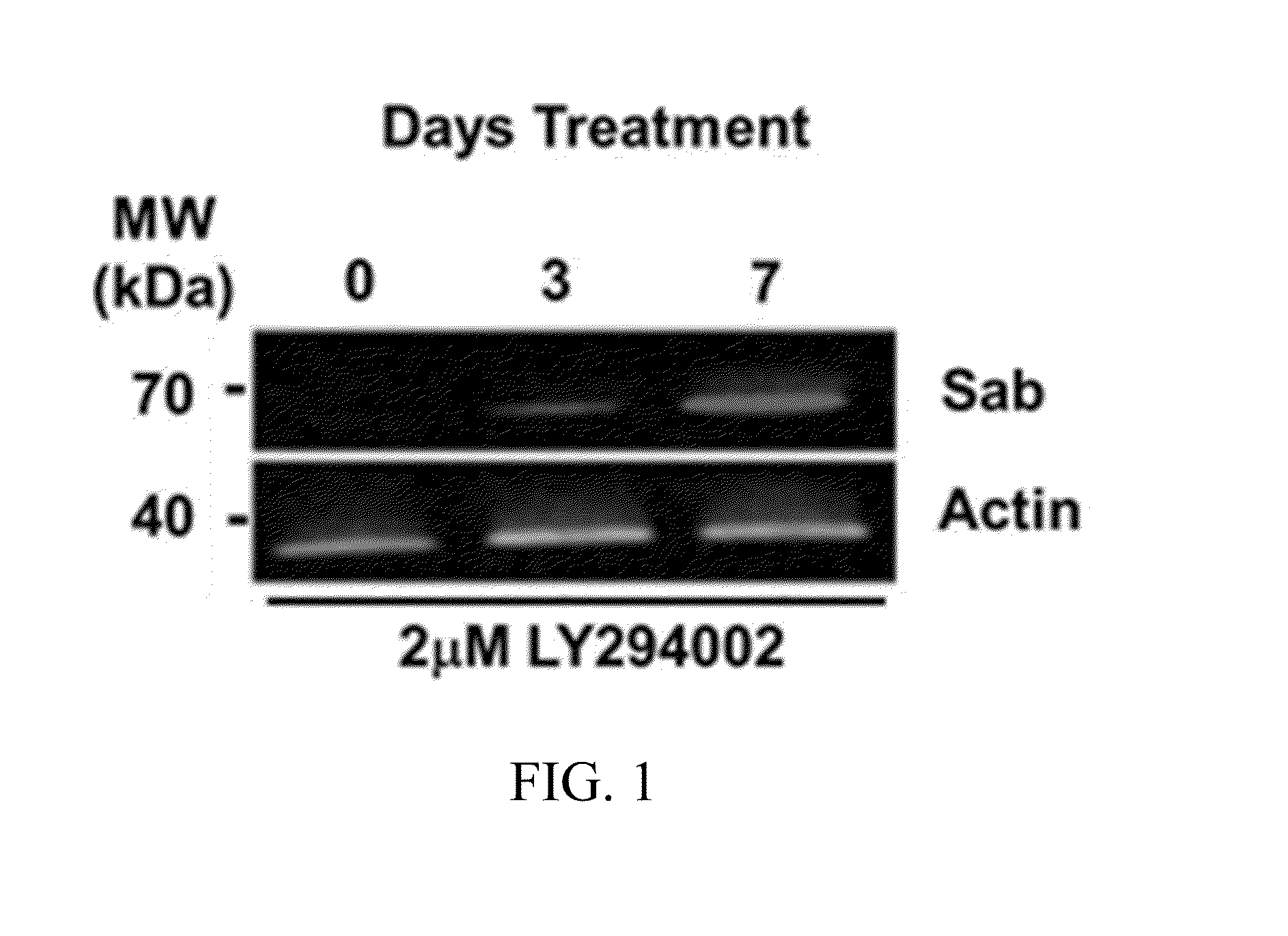
[0156]HeLa cell mitochondria were primed using a sub-chronic (2 μM dose) of...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Fluorescence | aaaaa | aaaaa |
| Toxicity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com