Glycoprotein biomarkers for esophageal adenocarcinoma and barrett's esophagus and uses thereof
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Identification of Biomarkers for EAC
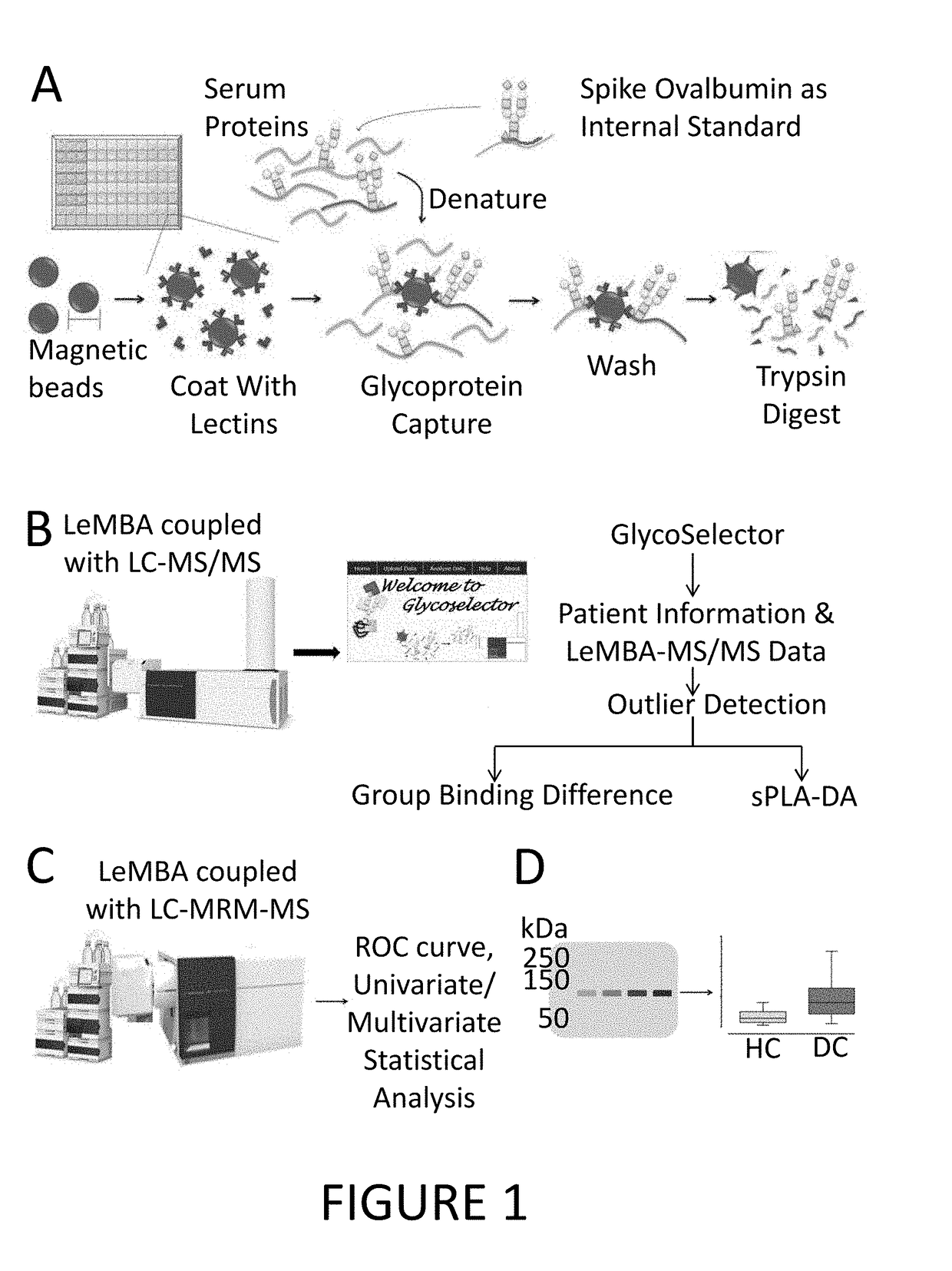
[0170]To identify biomarkers for EAC and BE, the abundance of proteins with altered glycosylation structures (i.e. different glycospecies) in the serum of healthy patients, patients with BE and patients with EAC was assessed using lectin-magnetic bead array-coupled mass spectrometry (LeMBA-MS) essentially as described by Choi et al. (Electrophoresis (2011) 32, 3564-3575). A schematic of the biomarker identification protocol is shown in FIG. 1.
Materials and Methods
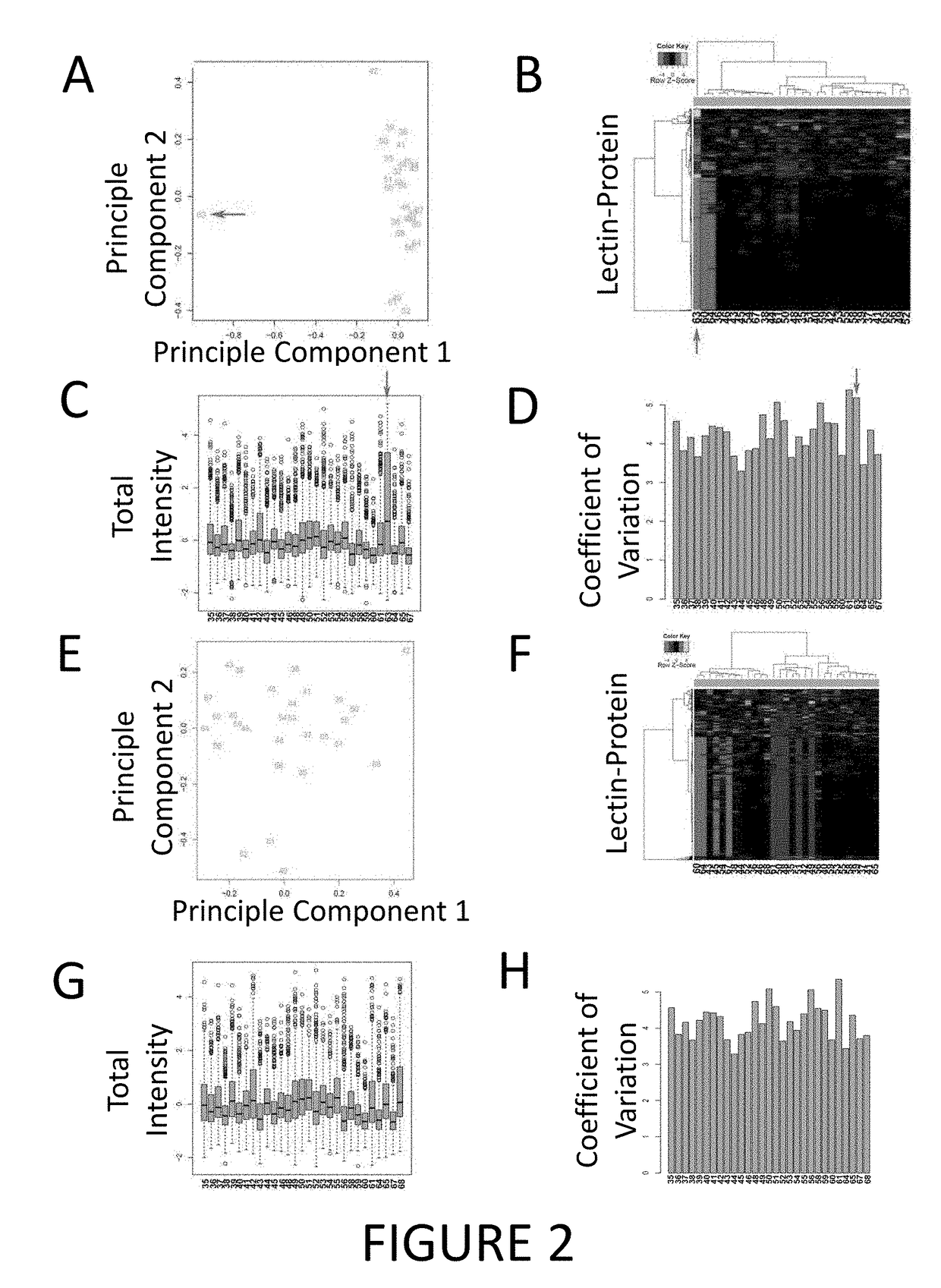
[0171]In the discovery phase, 29 serum samples (TABLE 8), consisting of 10 each of BE, EAC and 9 healthy controls (4 confirmed BE-free from Study of Digestive Health and 6 population controls from Australian Cancer Study), were analysed. One of the control patients subsequently developed BE, so the data were excluded from further analysis. All of the patients were male, reflecting the male-dominance of EAC and BE.
[0172]The serum samples were denatured by heating in denat...
example 2
Detecting Combinations of Glycospecies
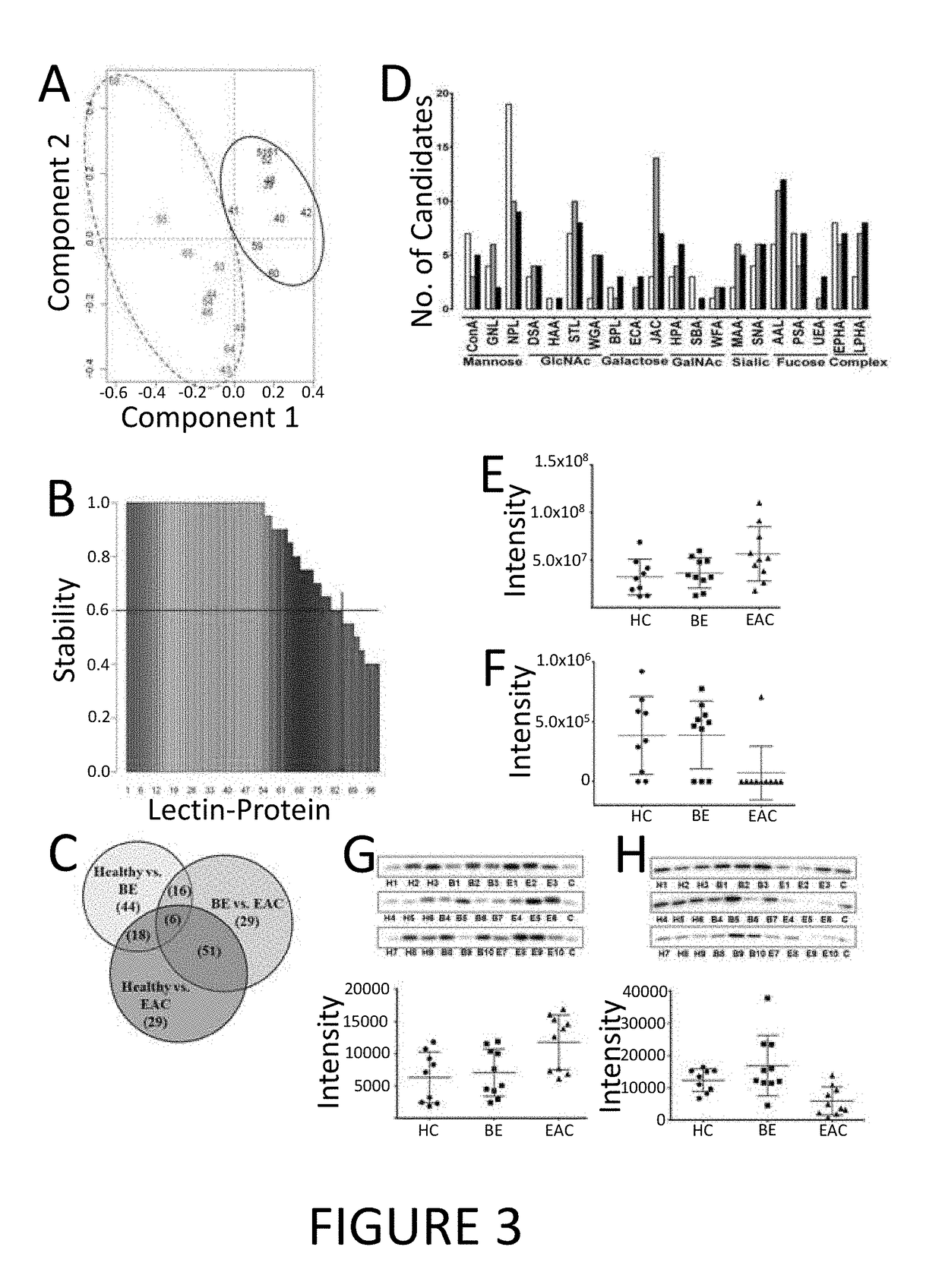
[0184]In order to determine whether a particularly robust set of markers could selected in order to improve the determination of the likelihood of the presence or absence of BE or EAC in a subject, combinations of glycospecies identified from the relevant table (TABLES 1, 4, and 6) were analysed.
[0185]It was found that the power of the diagnostic test could be enhanced by measuring a panel of two, three, four, five, or more than five markers (see, TABLE 11). Notably, when using four glycospecies (JAC-binding complement component C9, EPHA-binding alpha-1B-glycoprotein, EPHA-binding gelsolin, WGA-binding angiotensin and alpha-2-macroglobulin) are measured between subjects with EAC and healthy controls, an AUC of up to 98.25% can be achieved.
TABLE 11AUCGlycospecies (EAC v Healthy)P02748_JAC0.775P02748_JAC P04217_EPHA0.86P02748_JAC P04217_EPHA P06396_EPHA0.93P02748_JAC P04217_EPHA P06396_EPHA P01019_WGA0.9825P02748_JAC P04217_EPHA P06396_EPHA P01019...
example 3
Validation of Glycospecies Biomarkers
[0188]In order to demonstrate that glycospecies biomarkers can be reliably determine the likelihood of a subject having a relevant condition, a number of the biomarkers were selected for validation in a separate and distinct cohort of subjects.
[0189]TABLE 13 shows the relative increase or decrease of exemplary differentially glycosylated proteins in the serum of patients with EAC compared to healthy patients, BE compared to healthy patients, and in the serum of patients with EAC compared to patients with BE, i.e. the relative abundance of the specific glycospecies. For example, six proteins (P00738: haptoglobin, P00751: complement factor B, P01011: alpha-1-antichymotrypsin, P02748: complement component C9, P09871: complement Cls subcomponent, and P10643: complement component C7) having glycans that facilitated binding to EPHA (i.e. the EPHA-binding glycospecies of haptoglobin, the EPHA-binding glycospecies of complement factor B, the EPHA-binding...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com