Method for detecting mononucleotide polymorphism with biochip
A single nucleotide polymorphism, biochip technology, applied in biochemical equipment and methods, microbial determination/inspection, fluorescence/phosphorescence, etc., can solve the problems of high detection cost and difficulty in achieving high-throughput detection. Simplified operation steps, significant advantages, low non-specific binding rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
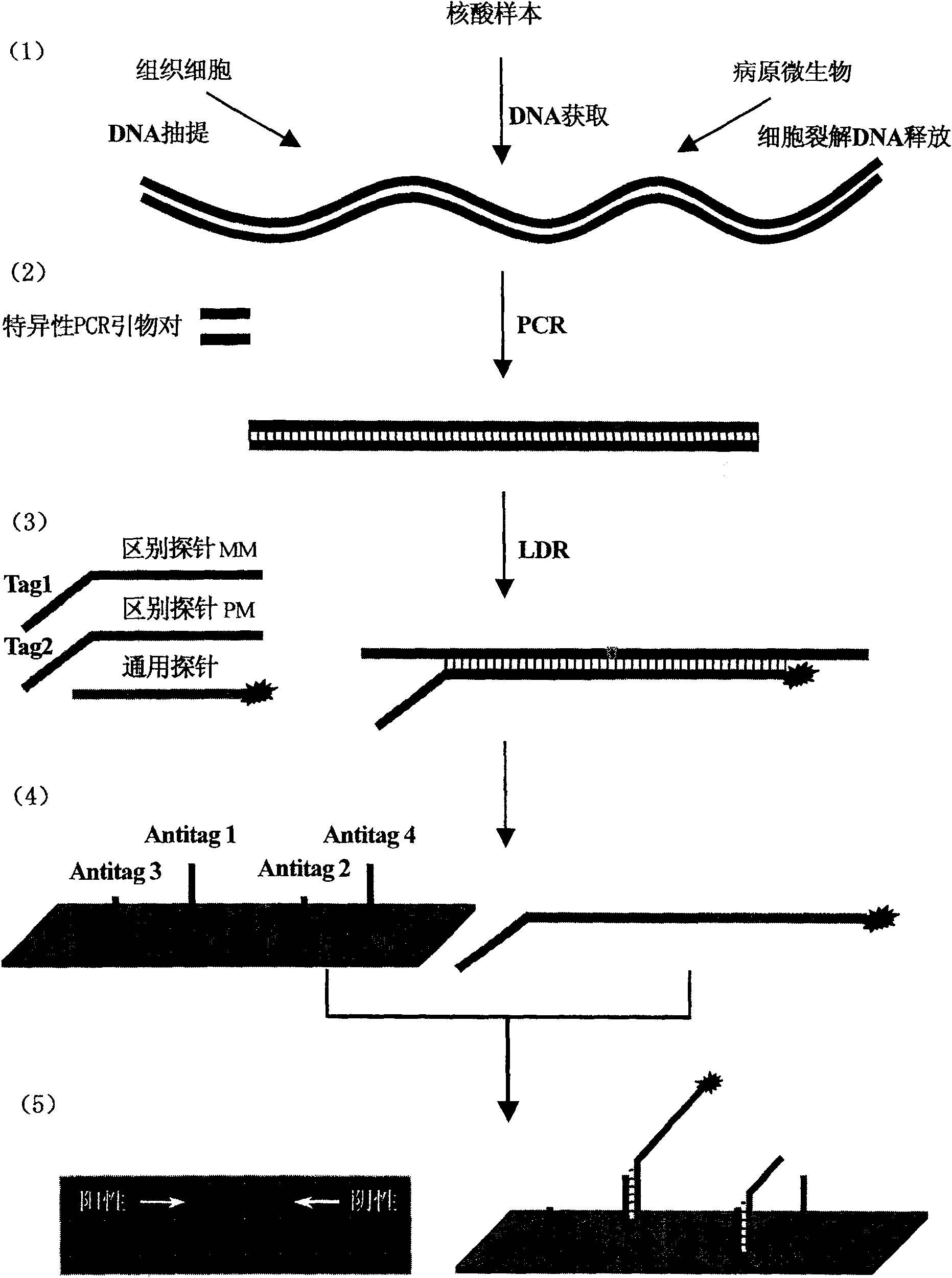
[0073] Nucleic acid amplification technology is a rapid and economical technique for synthesizing a large number of copies of DNA fragments. Commonly used in prenatal diagnosis of genetic diseases, detection and diagnosis of cancer genes, detection of pathogenic pathogens, DNA fingerprinting, individual identification, parent-child relationship identification and forensic evidence, animal and plant quarantine, rapid diagnostic testing, gene splicing, sequencing and other molecules field of biology.
[0074] The ligase reaction is a technique for linking two nucleic acid fragments, which is often used in the field of molecular biology such as molecular cloning.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com