Oncolytic adenoviruses for the treatment of cancer
An oncolytic adenovirus, adenovirus technology, applied in the field of tumor biology, can solve problems such as lack of potency, genome instability, and interference with promoter virus sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
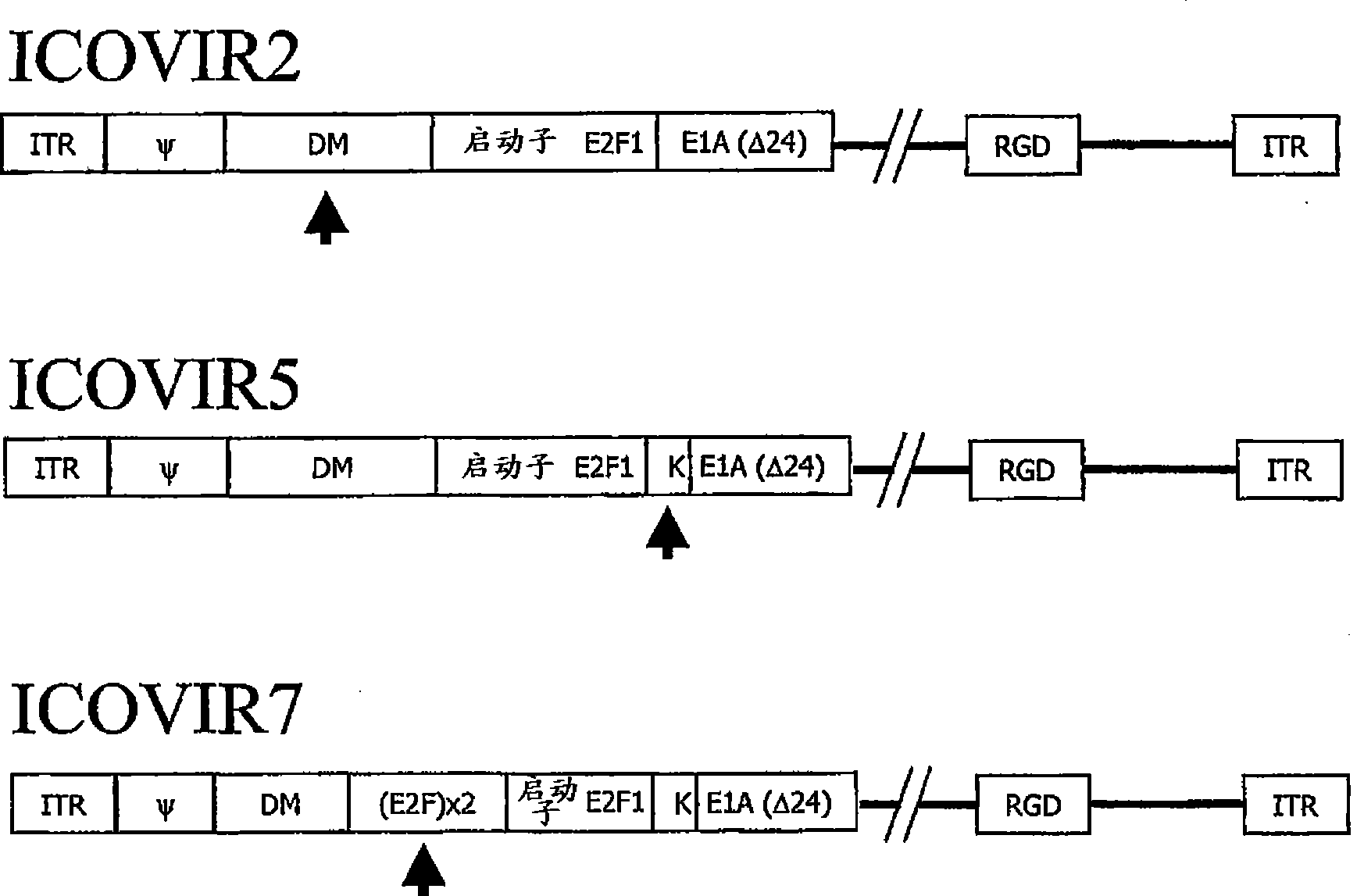
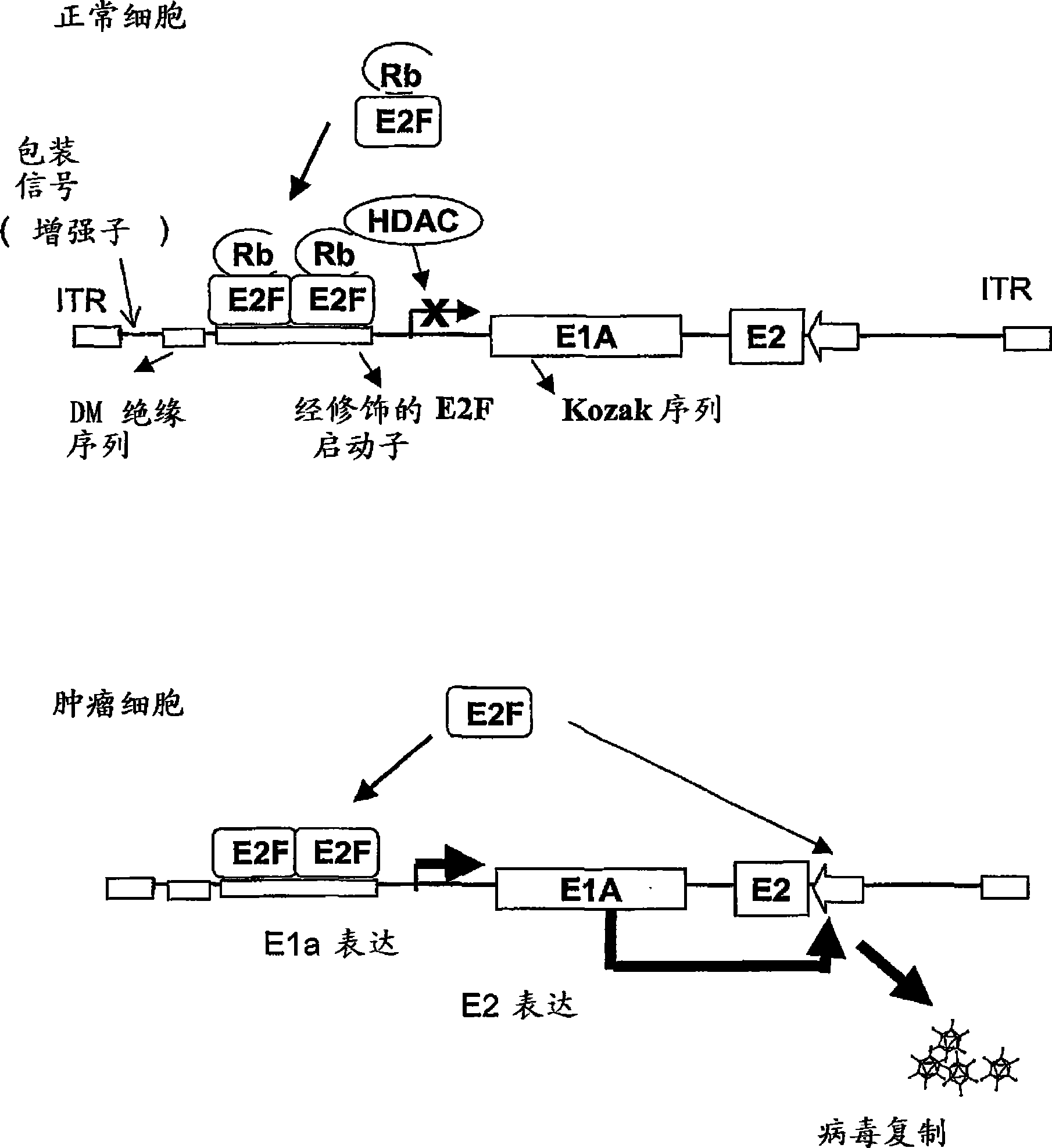
[0065] Oncolytic adenoviruses with E1a regulated by the E2F1 promoter separated by MD sequences express E1a and replicate selectively in tumor cells.
[0066] Adenoviruses with E1a regulated by the E2F1 promoter separated by MD sequences were constructed as follows: For the production of ICOVIR-1 (Ad-E2F-Δ24RGD), the human E2F1 promoter was obtained by PCR of human peripheral blood mononuclear cells using Oligonucleotides extending from -218 bp to +51 bp of the E2F1 promoter (position +1 indicates start of transcription). The oligonucleotides contained KpnI and HindIII restriction targets for cloning into plasmid pGL3 (Promega, Southampton, UK). The resulting plasmid was named pGL3-E2F. Thus, by removing (nt) 122 and 129 nucleotides of E1a (derived from pXC1-Δ24, which has a HindIII site between nt 348 and nt 522 of the Ad5 genome) 9 ) except for the 5766-base plasmid was recombined to obtain pE2F-Δ24. pE2F-Δ24 and pShuttle 41 Recombination was performed to obtain pShuttle...
Embodiment 2
[0070] The kozak sequence enhances E1a expression in oncolytic adenoviruses in which E1a is regulated by the E2F1 promoter separated by MD.
[0071]Oncolytic adenoviruses were constructed with E1a regulated by the E2F1 promoter separated by the MD sequence and a kozak sequence that enhances its translation. To this end, a DNA fragment containing the MD sequence, the E2F1 promoter and E1a was isolated from pShuttle-MD-E2F-D24 described in Example 1 by KpnI restriction digestion and subcloned into pGEM3Z (Promega) to obtain the plasmid pGEM- E2F-d24. This plasmid was used to replace the E1a translation origin with an oligonucleotide containing the kozak sequence to obtain pGEM-E2F-KD24. The thus modified KpnI fragment was recloned into KpnI of pShuttle-MD-E2F-D24 to obtain pShuttle-DM-E2F-KD24. Finally, pShuttle-MD-E2F-KD24 was recombined with pVK503 to obtain pICOVIR5. ICOVIR5 virus was generated by digesting this plasmid with PacI and transfected into HEK293 cells. ICOVIR5...
Embodiment 3
[0074] The kozak sequence confers increased oncolytic potency of adenovirus whose E1a expression is regulated by the E2F1 promoter separated by MD.
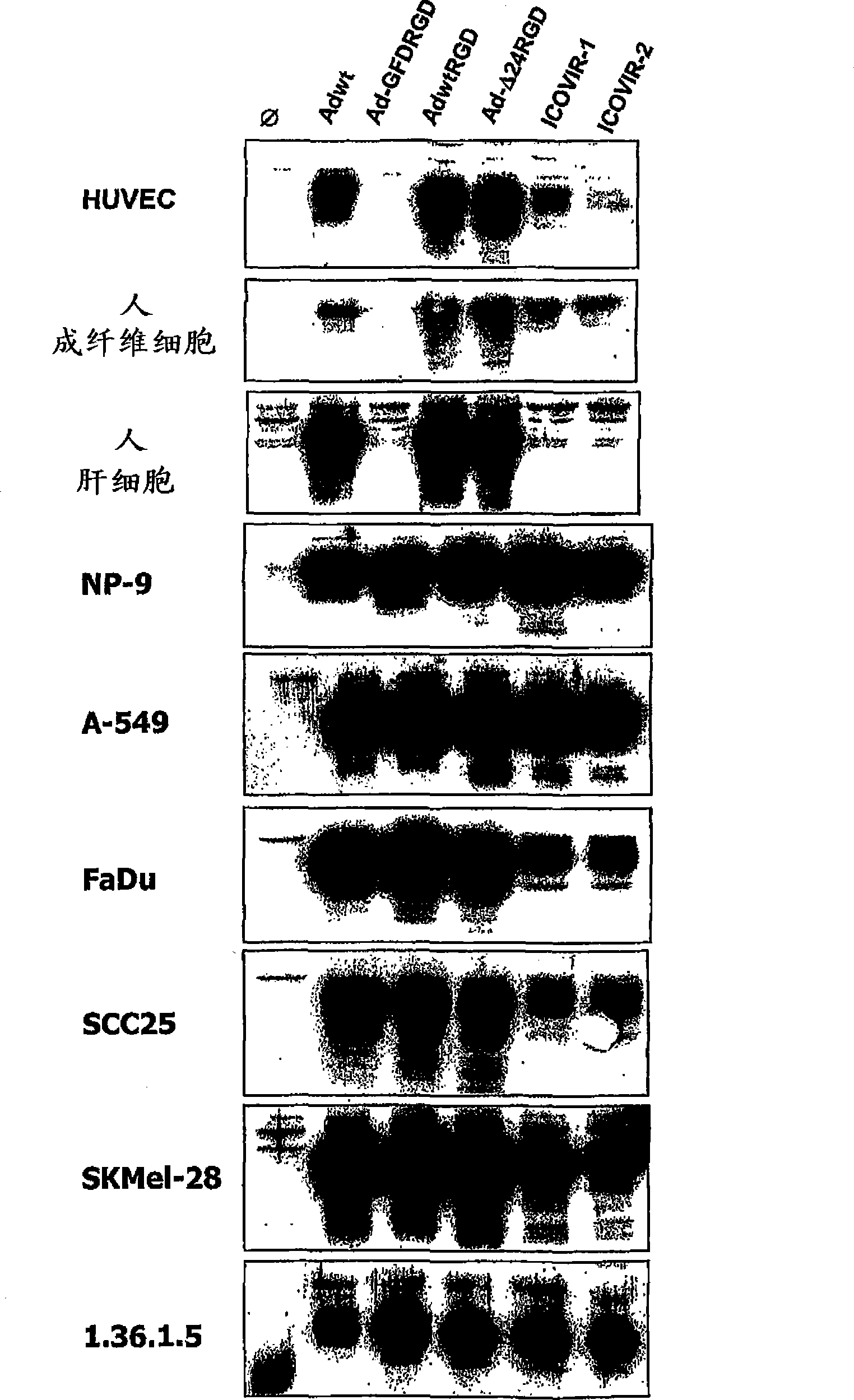
[0075] We cultured cells from the tumor cell lines SKMel-28 and FaDu in 96-well plates, where visible reduction of ICOVIR2 replication ability (as in Example 1 and Figure 4 described in ). These cells were infected with increasing amounts of ICOVIR5, ICOVIR2 and AdwtRGD (the latter serving as a control for maximal lytic capacity). Five days after infection, the amount of protein was measured spectroscopically as a reflection of cell survival. The results in the present invention Figure 6 displayed in . The cleavage ability of ICOVIR5 in SKMel-28 was the same as that of AdwtRGD and higher than that of ICOVIR2. In FaDu, it is also higher than ICOVIR2, although not to the level of AdwtRGD.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com