Method for assaying nucleic acids by fluorescence
一种荧光测定、核酸的技术,应用在荧光测定法领域,能够解决必需提取、高操作成本、灵敏性不足等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
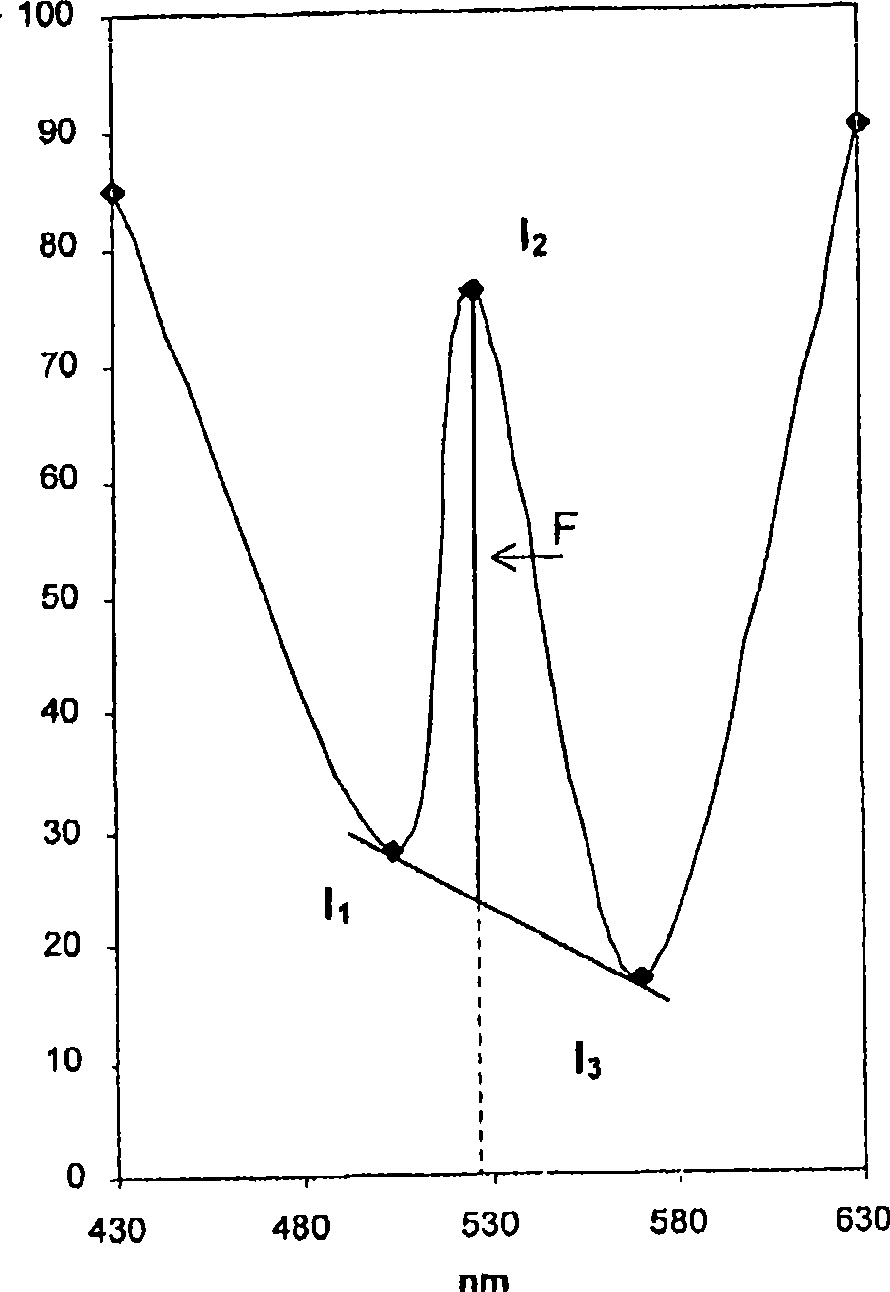
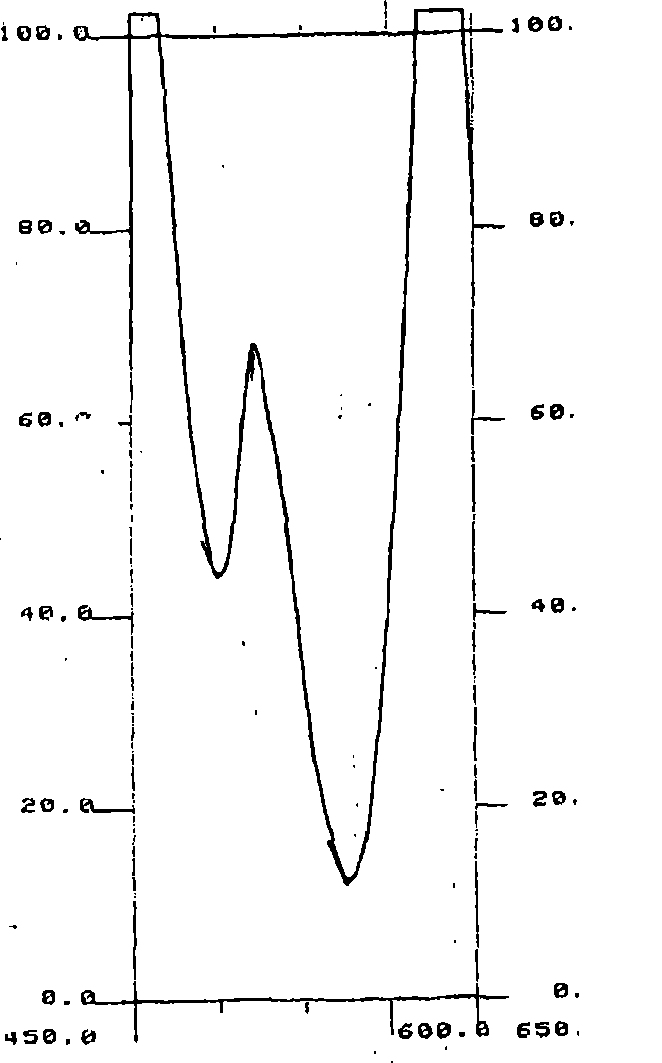
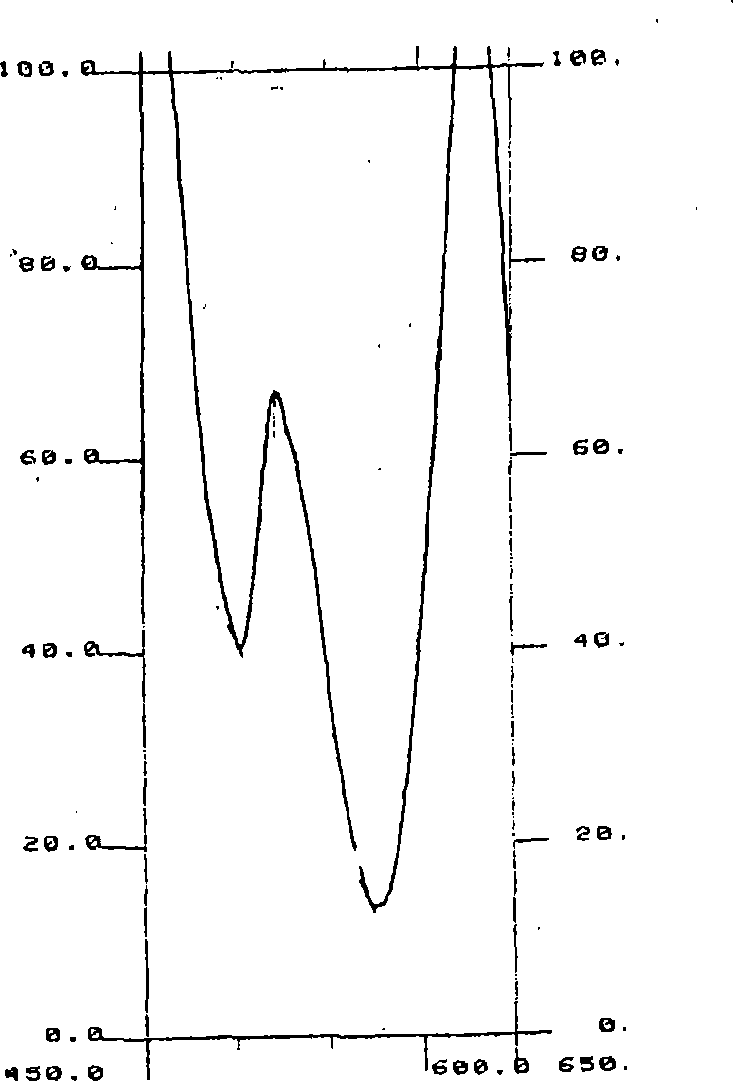
[0066] nucleic acid
[0067] Nucleic acids may be of natural or synthetic origin, and the nucleotides to which they are incorporated, especially ribonucleotides or deoxyribonucleotides, may be natural or modified. Preferably, the nucleic acid is DNA or RNA, more preferably DNA. Also preferably, the size of the nucleic acid strand is greater than five bases.
[0068] Advantageously, the specific quantification of nucleic acids in a sample with low molecular weight strands (e.g., less than 1,000 bases in length) (optionally after selecting these nucleic acids using a suitable method) allows quantification, for example, of the biological or cultural death due to apoptosis of cells in an organism; the value thus measured can be compared to the value of the total amount of DNA, optionally measured without the use of any method for selecting strands less than 1,000 base pairs in size correlated to provide the percentage of cell death due to apoptosis.
[0069] Apoptosis can also ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com