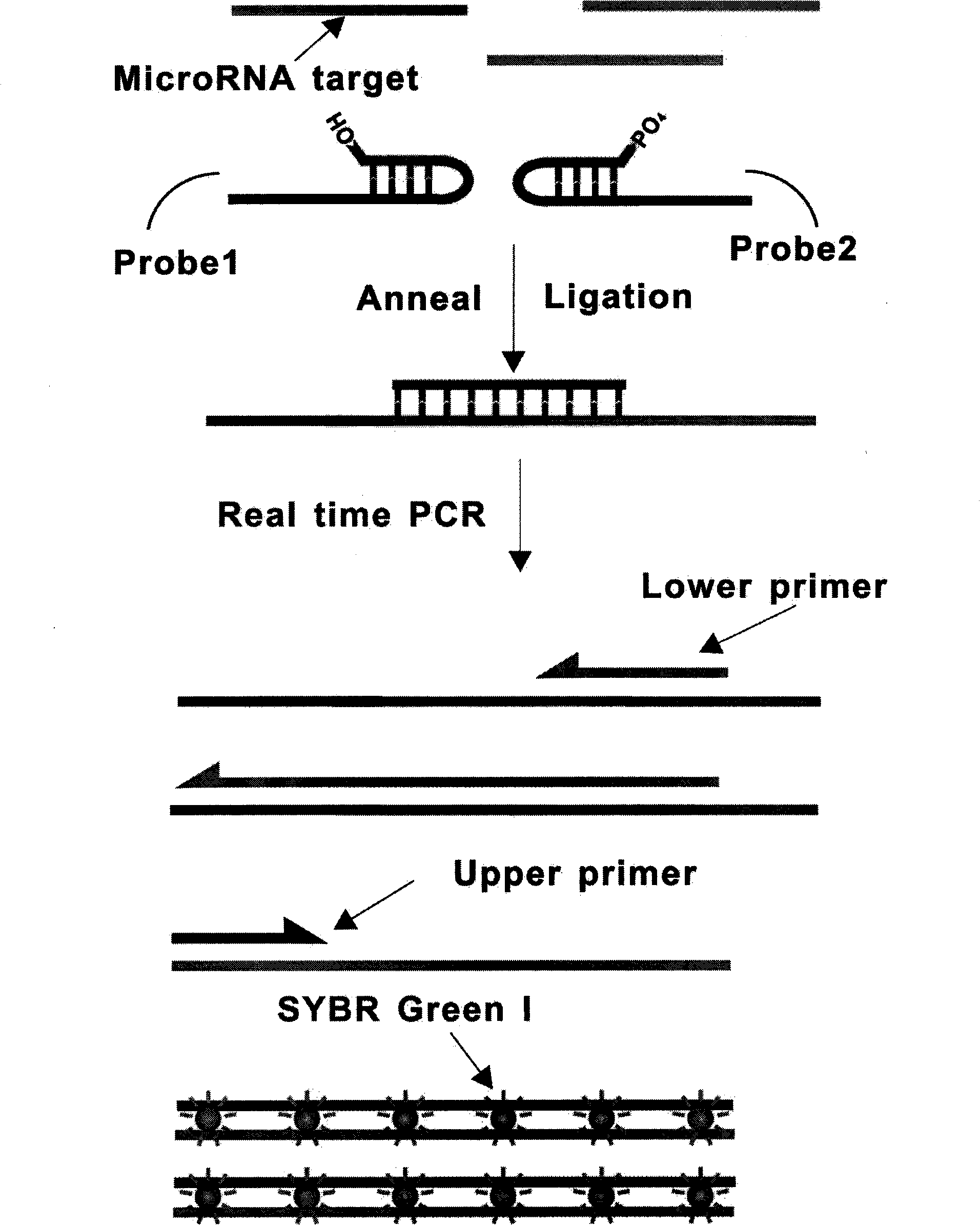
Probe set for detecting small RNA and method for detecting small RNA using same
A technology of DNA probes and probe sets, applied in the direction of DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of increasing the difficulty of preparation and the cost of experiments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
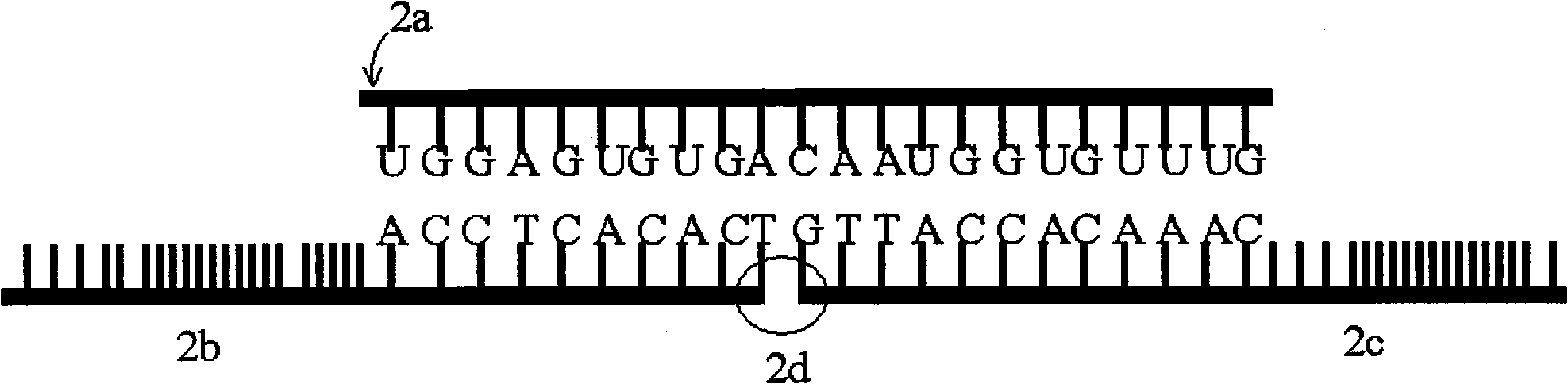
[0037] Design and acquisition of linear probes. The following will combine figure 2 The design of the linear probe is described in detail. figure 2 2a in the figure is a schematic diagram of miRNA-122 sequence, which has 22 nucleotides and is not long enough for PCR amplification. Probe 2b has 31 nucleotides, and 2c has 30 nucleotides. When the pair of DNA probes are first hybridized with miRNA-122 and then connected at 2d under the action of T4 DNA ligase, they can A DNA strand of 61 nucleotides is formed, which is long enough for PCR amplification. The DNA probes used in the experiment were purchased from Invitrogen Company in Shanghai, China.
Embodiment 2
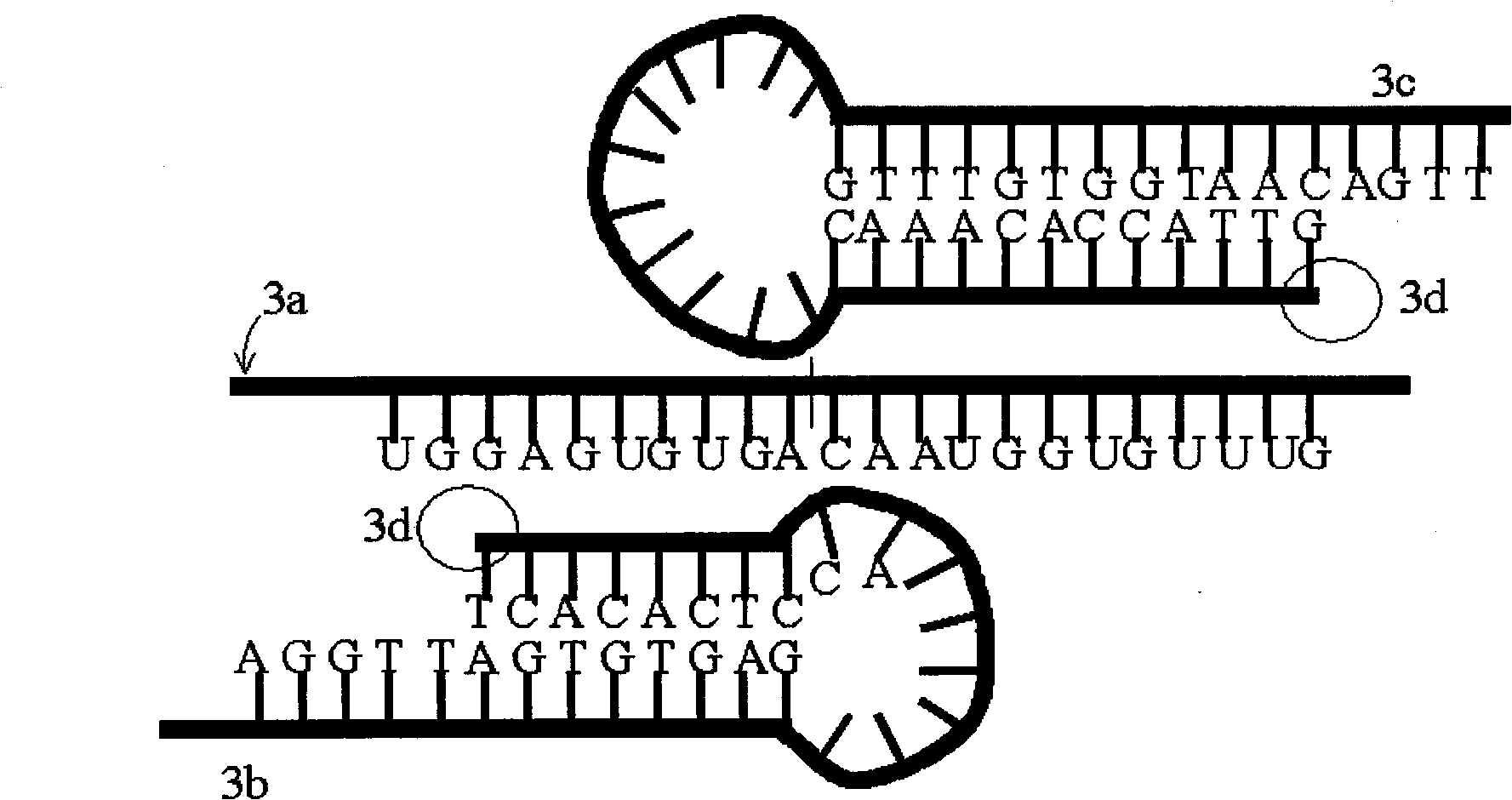
[0039] Design of probes with a stem-loop structure. The following will combine image 3 The design of a probe with a stem-loop structure is detailed. image 3 3a in the figure is a schematic diagram of miRNA-122 sequence, which has 22 nucleotides and is not long enough for PCR amplification. Probe 3b has 33 nucleotides, and 3c has 39 nucleotides. When the temperature rises, the stem-loop structure of this pair of probes can be opened to become a straight-chain DNA molecule, and then hybridized with miRNA-122. Then, under the action of T4 DNA ligase, after joining at 3d, a DNA chain with 72 nucleotides can be formed, which is long enough for PCR amplification.
Embodiment 3
[0041] The detection method of miRNA. In this example, a probe with a stem-loop structure similar to that in Example 2 will be used to detect target miRNAs from a system containing 6 miRNAs. The six miRNAs are Mmu-mir-122, syn-aa, aga-mir-210, has-mir-549, has-mir-214 and has-mir-21, and their nucleotide sequences are shown in Figure 4 and shown in Table 1. Mmu-mir-122 is the target miRNA. Among the six miRNAs, except for the syn-aa sequence which was artificially designed to meet the requirements of this experiment, the rest of the miRNA sequences were obtained from the Sanger database. The above six miRNAs used in the experiment were all purchased from Shanghai Gemma Pharmaceutical Technology Co., Ltd. Figure 4 The part of the nucleotide sequence identical to the target miRNA in the five non-target miRNA nucleotide sequences is shown with an emphasis number. It can be seen from the figure that there are 2-8 nucleotide sequences in the non-target miRNA that are identical...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com