Methods of detecting long range chromosomal interactions
A long-distance, chromosomal technology, used in biochemical equipment and methods, pharmaceutical formulations, and microbial determination/inspection, which can solve problems such as histone acetylation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] Example 1: Epigenetic control of the GAL locus
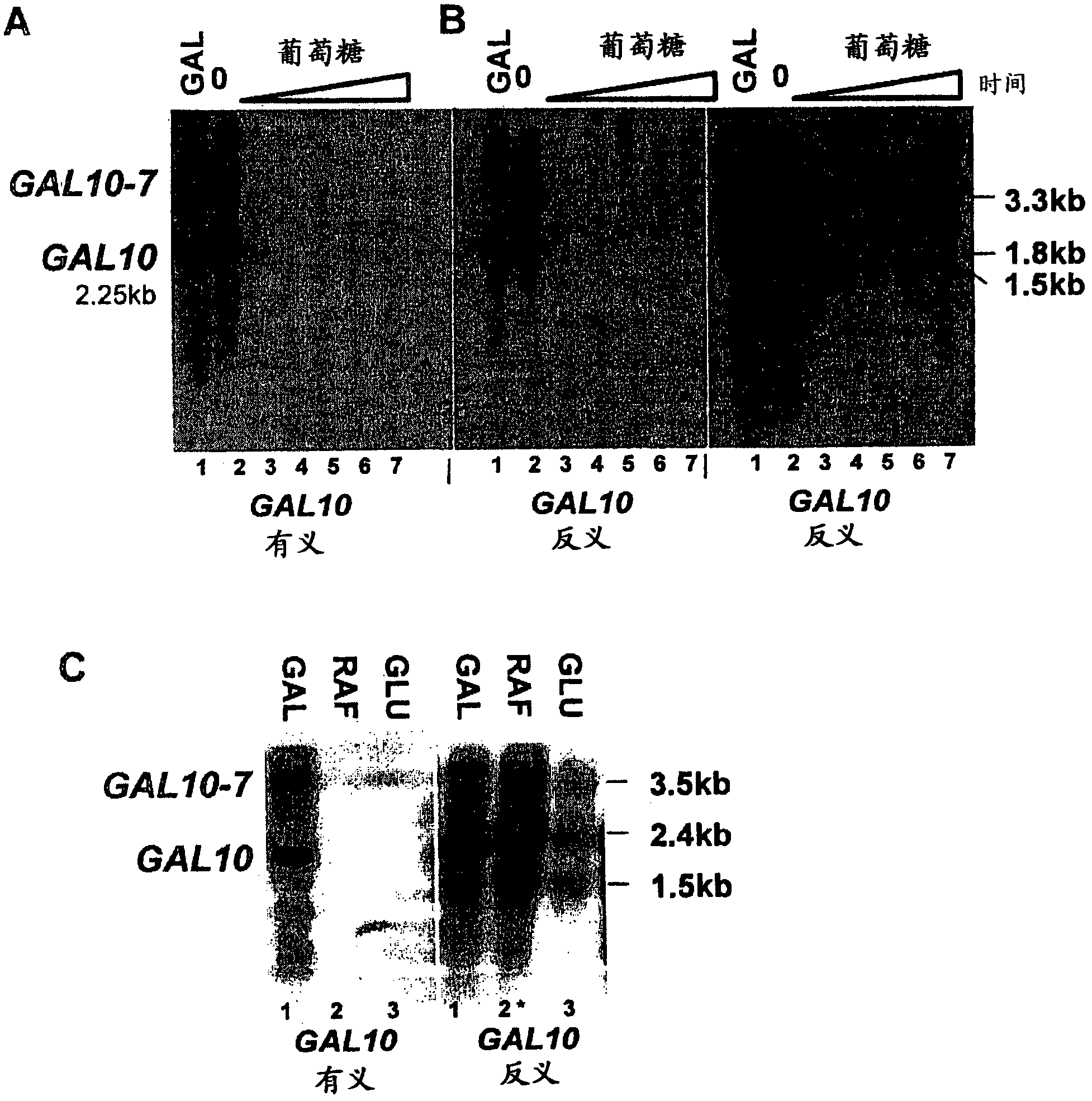
[0093] Conditional antisense transcript at GAL10.
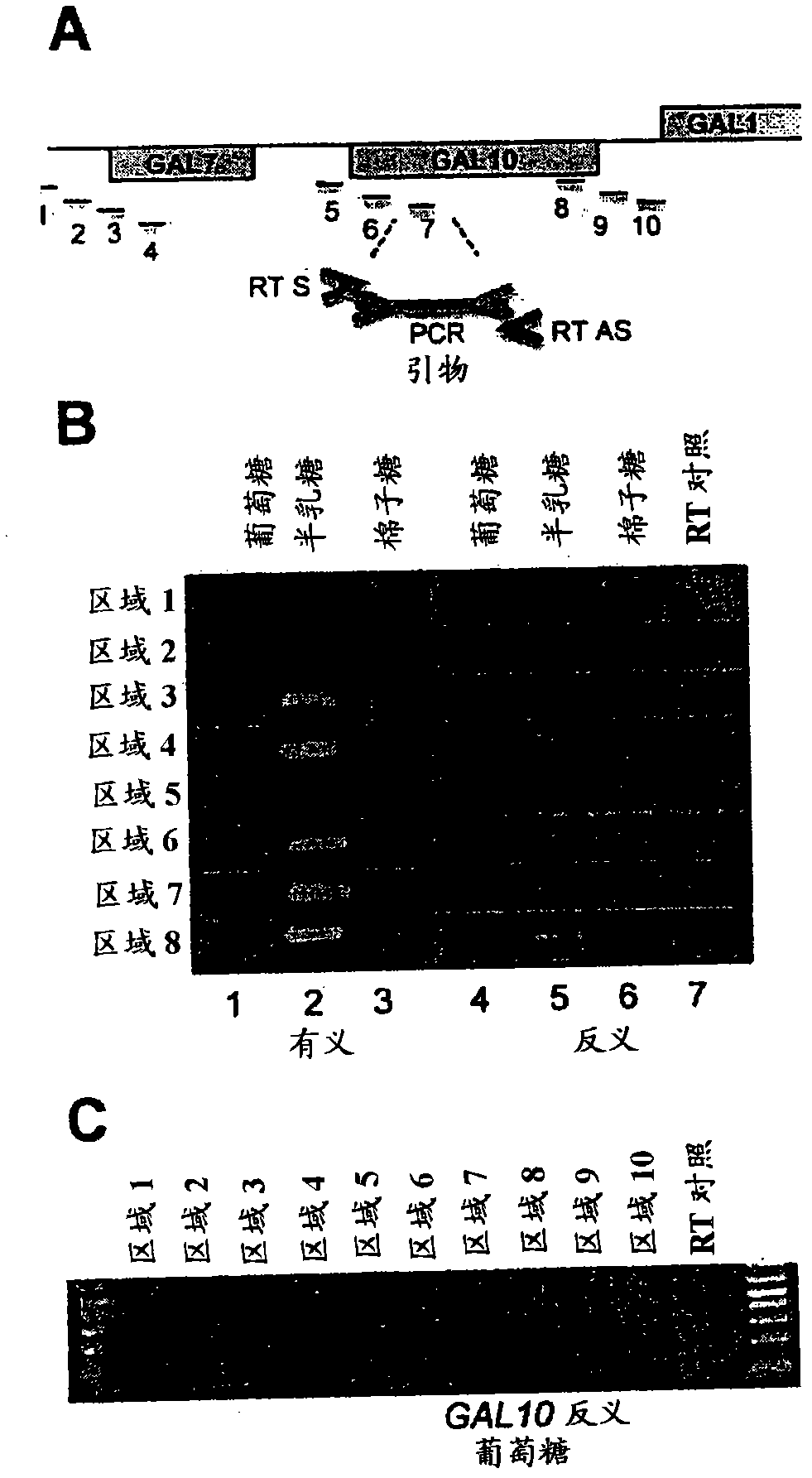
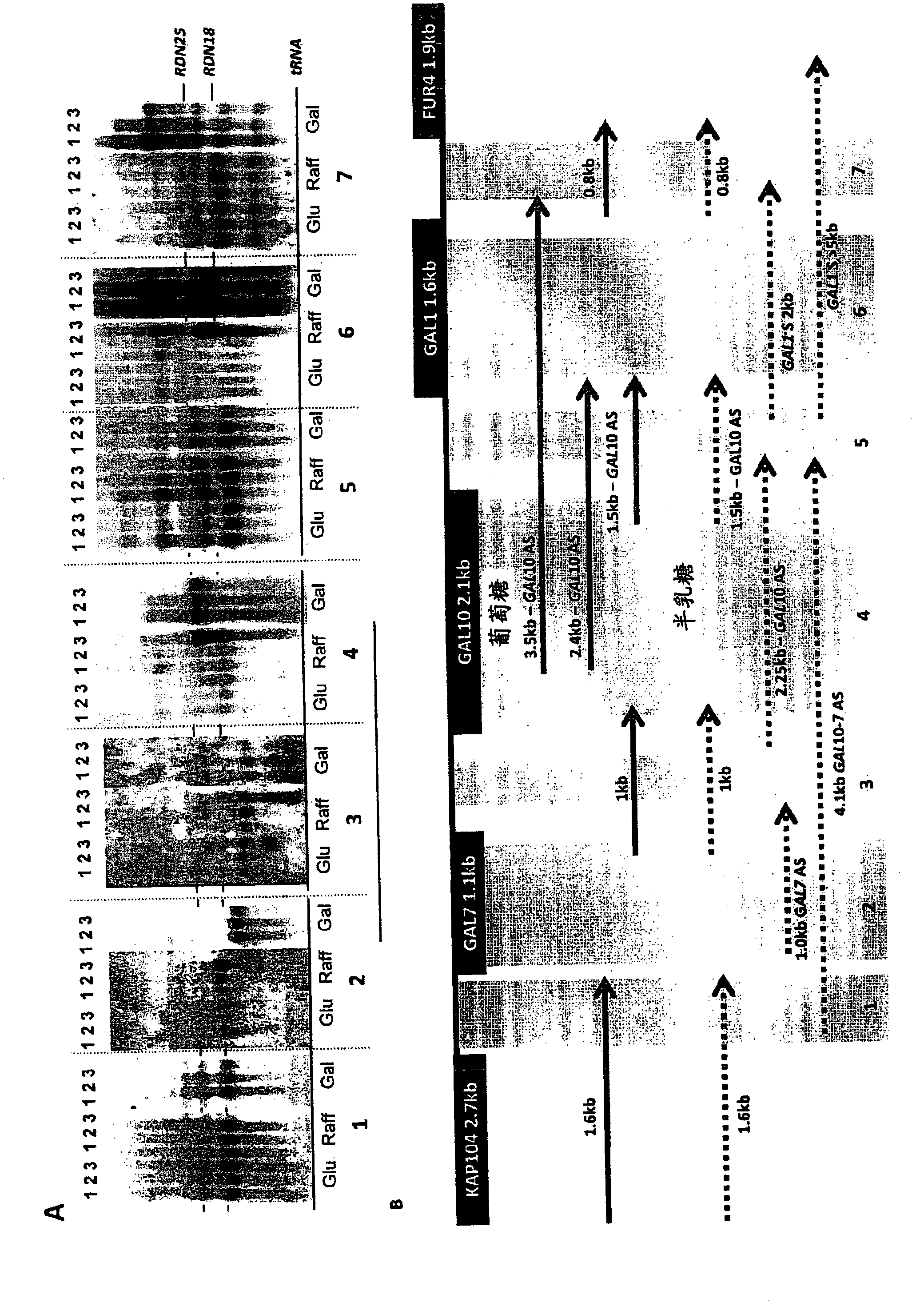
[0094] Addition of glucose to cultures of cells grown in galactose resulted in a rapid repression of transcription. As expected, within 15 minutes of glucose addition, the 2.25 kb GAL10 transcript and the longer 4.1 kb GAL10-7 fusion transcript (Greger and Proudfoot, 1998, Embo J 17, 4771-4779; St. John and Davis, 1981 , J Mol Biol 152, 285-315) the levels of both decreased significantly ( figure 1 A). Antisense transcripts have been reported at the repressed GAL10 gene (Perocchi et al, 2007, Nucleic Acids Res 35, e128; Samanta et al, 2006, Proc Natl Acad Sci USA 103, 4192-4197; David et al, 2006 , Proc Natl Acad Sci USA 103, 5320-5325; Miura et al, 2006, PNAS 103, 17846-17851). The inventors investigated whether antisense transcripts are a general feature of the GAL10 gene and, if so, when these transcripts arise.
[0095] In induced cultures, three antisense trans...
Embodiment 2
[0241] Example 2: Control of long-range interactions by CTCF
[0242] Several transcription factors have been shown to play a role in long-range DNA interactions and transcription and thus may be good candidates to provide a link between these processes. Examples include the essential transcription factor TFIIB, which has been shown to organize the looping of several genes in yeast (Mol Cell, 27, 806-16); and the transcription factors EKLF, GATA-1 and FOG-1, which are responsible for β-globin Long-range DNA interactions in genes (Drissen et al, 2004, Genes Dev, 18, 2485-90; Vakoc, et al, 2005, Mol Cell, 17, 453-62).
[0243] The inventors identified the CCCTC-binding protein (CTCF) as another candidate for carrying out these functions throughout the genome. CTCF is involved in the regulation of transcription and the formation of higher-order conformational intrachromosomal and interchromosomal structures (Klenova, 2002, Semin Cancer Biol, 12, 399-414; Kurukuti et al, 2006, Pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com