Kit and method for identifying and detecting Cordyceps sinensis
A Cordyceps sinensis and detection method technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, fluorescence/phosphorescence, etc., can solve the problems of easy pollution, detection speed and efficiency constraints, and quantitative detection. Less, accurate and effective test results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
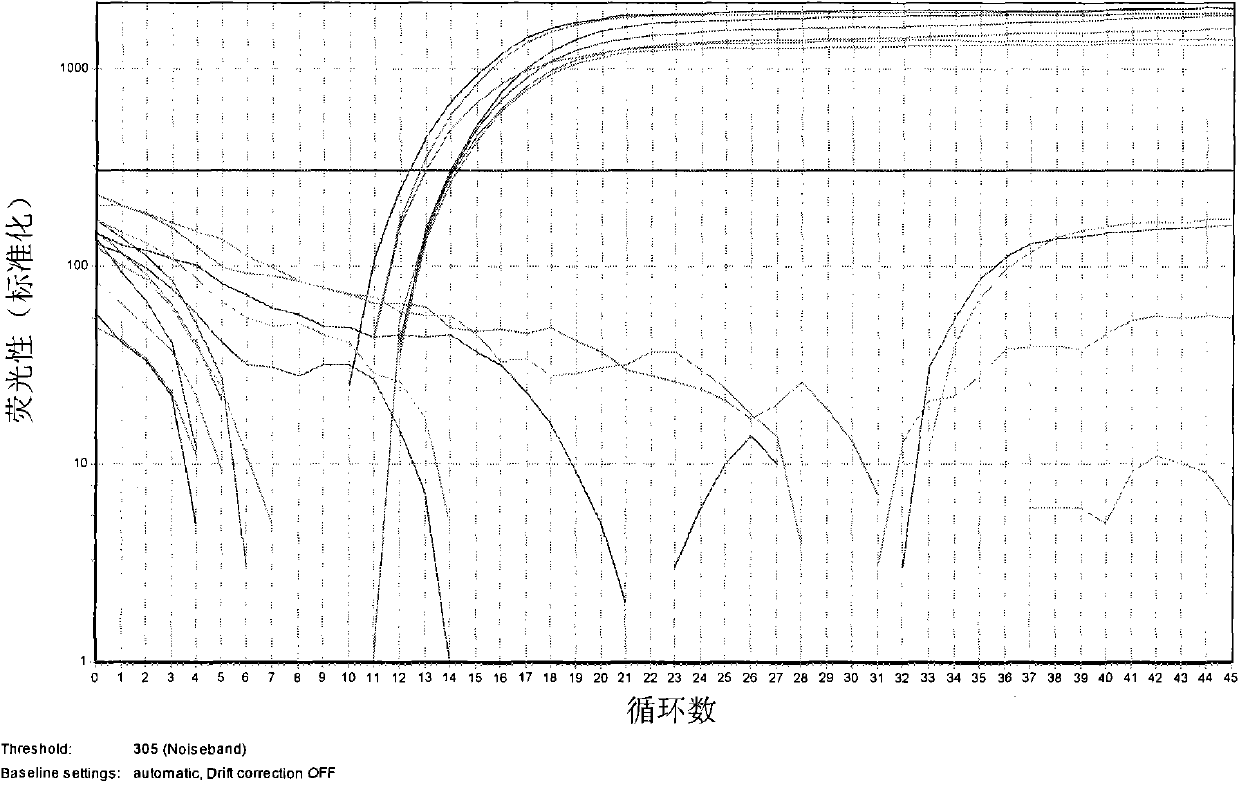
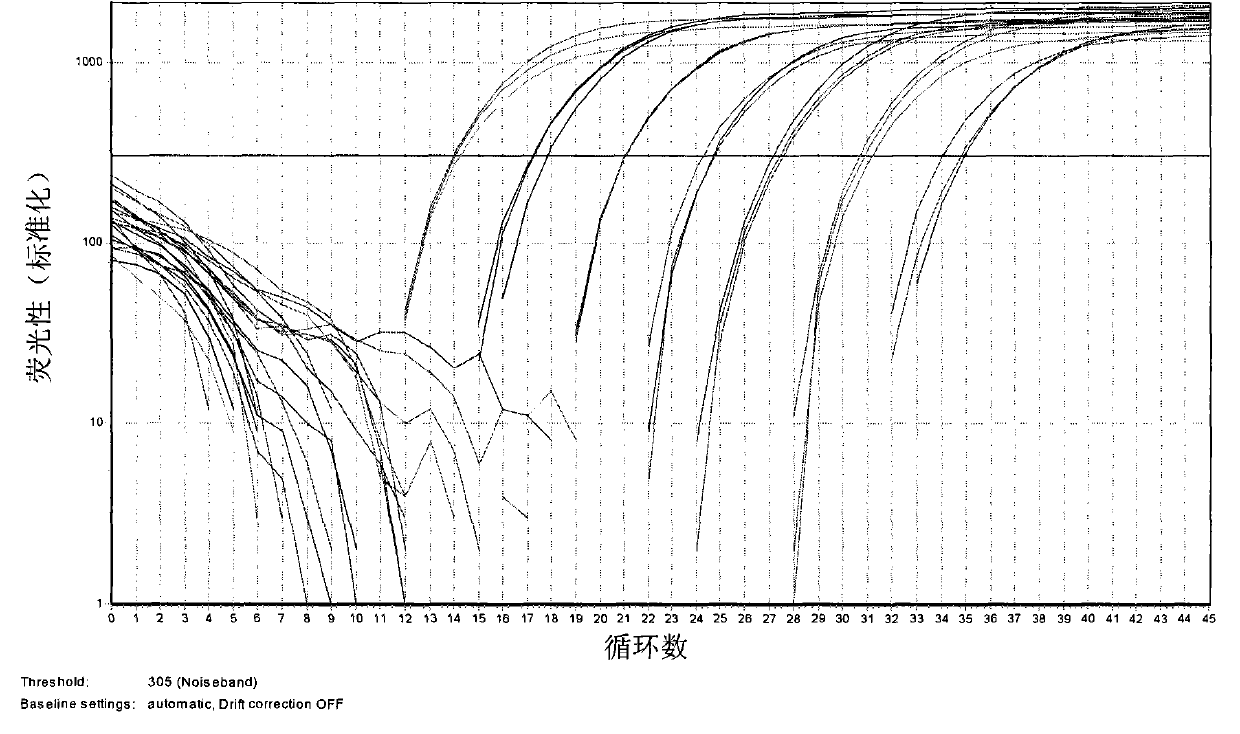
[0029] 1. Accuracy test - specific amplification of natural Cordyceps sinensis
[0030] Test procedure: using methods well known to those skilled in the art to extract natural Cordyceps sinensis (7 samples of natural Cordyceps sinensis collected from different villages and towns in Nyingchi, Tibet), Cordyceps militaris, Cordyceps guni, Cordyceps yunnanensis 1 and 2 (cordyceps spp. .) ribosomal DNA (rDNA), then with the primer pair and fluorescent probe provided by the present invention (sense primer is SEQ ID NO.1:5'-CAAGGTCTCCGTTGGTGA-3', antisense primer is SEQ ID NO.2: 5'-AACTGCTGTGGTGTTC C-3', the fluorescent probe is SEQ ID NO.3: 5'-Fam-CGGAGGGATCATTATCGAGTYACCACT-Tamra-3') for real-time fluorescent PCR, obtained as figure 1 The resulting figure is shown.
[0031] 1. Extraction of ribosomal DNA from the sample to be tested
[0032]As an example, the method for extracting ribosomal DNA of the present invention is as follows: using a Promega kit (article number: A1120),...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com