Sequencing method for clinical detection
A technology for sequencing and gene detection, applied in fluorescence/phosphorescence, material excitation analysis, etc., can solve the problems of missed detection of low-concentration templates, loss of PCR products, long operation time, etc., to avoid the generation of aerosols and reduce false positives. , the effect of reducing the missed detection rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1: Design of primers and probes.
[0036] For the highly mutated region of the P gene of hepatitis B virus, design specific PCR primers, probes and sequencing primers, the sequences of which are shown in the table below, and entrust a professional company to perform synthesis and fluorescent labeling.
[0037] name
Embodiment 2
[0038] Example 2: DNA template preparation.
[0039] Genomic DNA of hepatitis B virus was extracted from human serum using a viral nucleic acid extraction kit (QIAamp MinElute Virus Spin Kit, QIAGEN).
Embodiment 3
[0040] Example 3: PCR reaction.
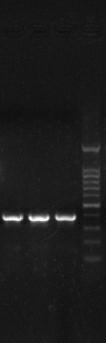
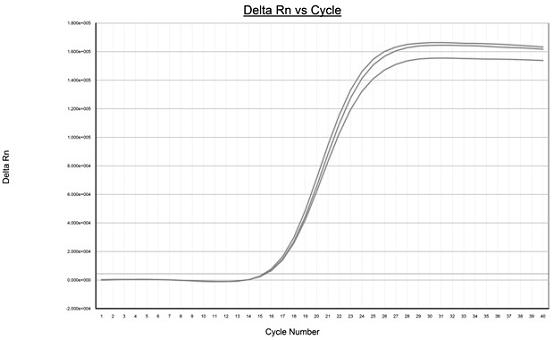
[0041] Use ABI7500 type fluorescent quantitative PCR instrument to carry out fluorescent quantitative PCR reaction on the highly mutated region of hepatitis B virus P gene, the results are as follows figure 2 As shown, the PCR products were detected by electrophoresis at the same time, and the results were as follows image 3 shown.
[0042] The reaction system of fluorescent quantitative PCR is:
[0043] DNA template: the hepatitis B virus genomic DNA sample obtained in Example 1, 5 μl;
[0044] 0.4 μmol / L each of upstream and downstream PCR primers (final concentration);
[0045] Probe 0.12 μmol / L (final concentration);
[0046] TaqMan Gene Expression Master Mix (including PCR reaction solution, high temperature-resistant DNA polymerase, uracil-N-glycosylase, etc.) from American LifeTech Company, 12.5 μl;
[0047] Make up to 25 μl with ultrapure water.
[0048] The cycle parameters of fluorescent quantitative PCR are:
[0049] 37°C, ...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap