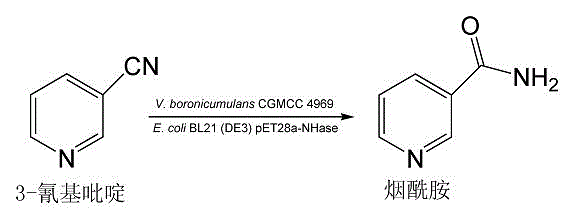
Variovoraxboronicumulans CGMCC 4969 and use thereof in bioconversion of 3-cyanopyridine for forming nicotinamide
A technology of biotransformation and greedy phagocytosis, applied in the field of microorganisms, can solve problems such as the function of nitrile hydratase that has not been experimentally proved
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
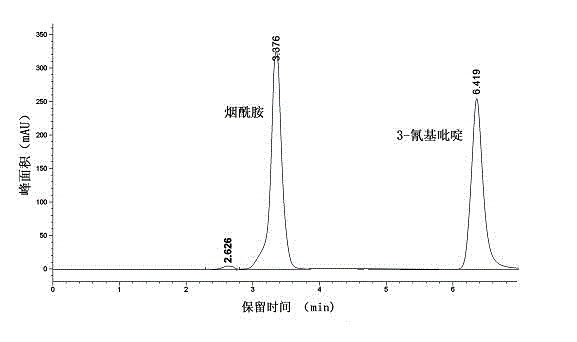
[0016] Example 1: Isolation, screening, identification and biological characteristics of strains that can biotransform 3-cyanopyridine into nicotinamide
[0017] 1. Strain isolation
[0018] Collect soil from Xianlin area, Nanjing City, Jiangsu Province, take 1g of soil and add it to 19mL sterile water containing 5 glass beads, shake it for 5min, let it stand for 10min, take 1ml of the suspension and add 19ml of mineral containing 1% 3-cyanopyridine in salt medium. The composition of the mineral salt medium is: 1.36 g / L KH 2 PO 4 , 2.13g / L Na 2 HPO 4 , 0.5 g / L MgSO 4 ·7H 2 O and 10ml / L metal ionic liquid, pH 7.5. The metal ionic liquid composition is: 0.40 g / L CaCl 2 2H 2 O, 0.30 g / L H 3 BO 3 , 0.04 g / L CuSO 4 ·5H 2 O, 0.10 g / L KI, 0.20 g / L FeSO 4 ·7H 2 O, 0.40 g / L MnSO 4 ·7H 2 O, 0.20 g / L NaMoO 4 2H 2 O and 10.0 mL / L concentrated hydrochloric acid. The samples were incubated at 30°C for 3 days in a shaker with a rotational speed of 220 rpm. Take 100 μ...
example 2
[0024] Example two: V. boronicumulans Cloning of CGMCC 4969 Nitrile Hydratase Gene
[0025] V. boronicumulans The extraction of genomic DNA of CGMCC 4969 is the same as the extraction method of genomic DNA in Example 1.
[0026] The four nucleotides involved are: deoxyadenine triphosphate (abbreviated as A), deoxythymidine triphosphate (denoted as T), deoxyguanine triphosphate (abbreviated as G) , Deoxycytosine nucleotide triphosphate (abbreviated as C). Design and synthesize the upstream and downstream primers of nitrile hydratase gene. R in the primer indicates that the base at this position is A or G, S in the primer indicates that the base at this position is C or G, Y in the primer indicates that the base at this position is C or T, and K in the primer indicates the base at this position The base is G or T.
[0027] Cloning with degenerate primers V. boronicumulans CGMCC 4969 nitrile hydratase alpha subunit gene fragment. The sequence of the synthesized pri...
example 3
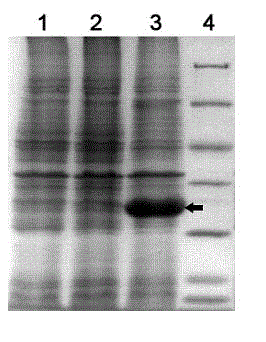
[0032] Example three: expression of nitrile hydratase gene and gene cluster containing recombinant nitrile hydratase E. coli For the biotransformation of 3-cyanopyridine
[0033] 1. The nitrile hydratase gene DNA fragment was connected to the plasmid pET28a
[0034] The primer NHCo-E-f containing the EcoRI restriction site and the primer NHCo-E-r containing the XhoI restriction site were designed. The sequence of the primer NHCo-E-f consists of 28 nucleotide residues, which are: 5′- GGGGAATTCATGACCGGCCATGACCACT-3 ' (SEQ ID No: 13); the sequence of the primer NHCo-E-r consists of 27 nucleotide residues, which are: 5'-GGGCTCGAGTGCCGCG GGCTCCAGGTA-3' (SEQ ID No: 14). In a sterilized 0.2 mL PCR thin-walled tube, add 10.8 μL sterile water, 2 μL amplification buffer, 2 μL four deoxynucleotides, 0.5 μL primer one, 0.5 μL primer two, 3 μL Genomic DNA solution prepared in step 1, 1 μL of DMSO, 0.2 μL of Angel DNA polymerase, the total volume is 20 μL; put the PCR tube in the PCR...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap