Method for screening micro ribonucleic acid (microRNA) target gene based on natural language processing system
A technology of natural language processing and screening method, applied in the field of bioinformatics screening of microRNA target genes, can solve problems such as target gene verification obstacles, and achieve the effect of easy identification.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
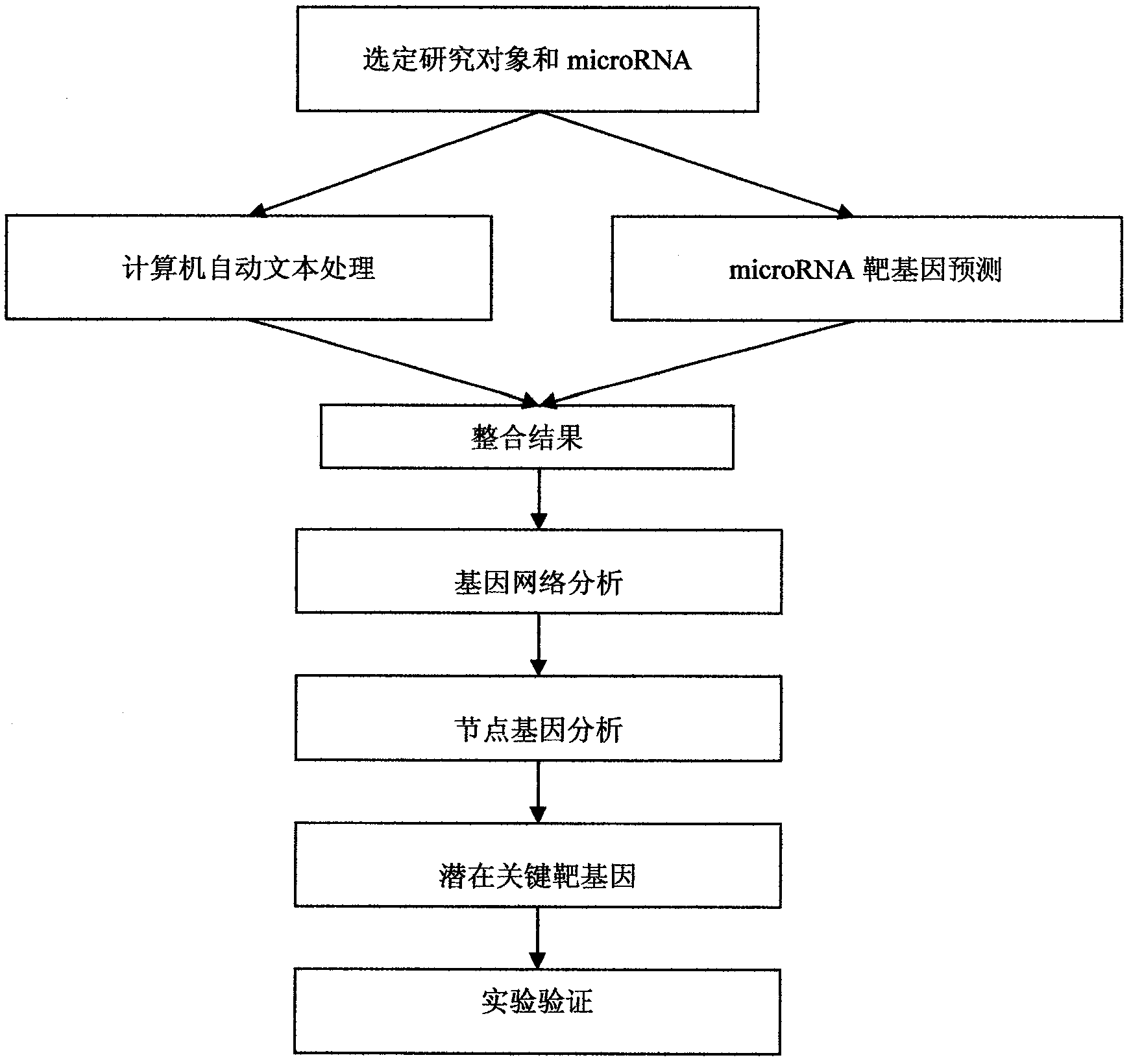
[0018] The present invention will take the target gene screening of miR-21 (a known microRNA) related to the occurrence of rectal cancer as an example to introduce the specific implementation steps of the method of the present invention.
[0019] Step 1. In the present invention, we choose the biological process of rectal cancer as the research object. By studying its tumor cells, we find that miR-21 is related to the formation of tumors, so we choose miR-21 as the microRNA for our experimental analysis. .
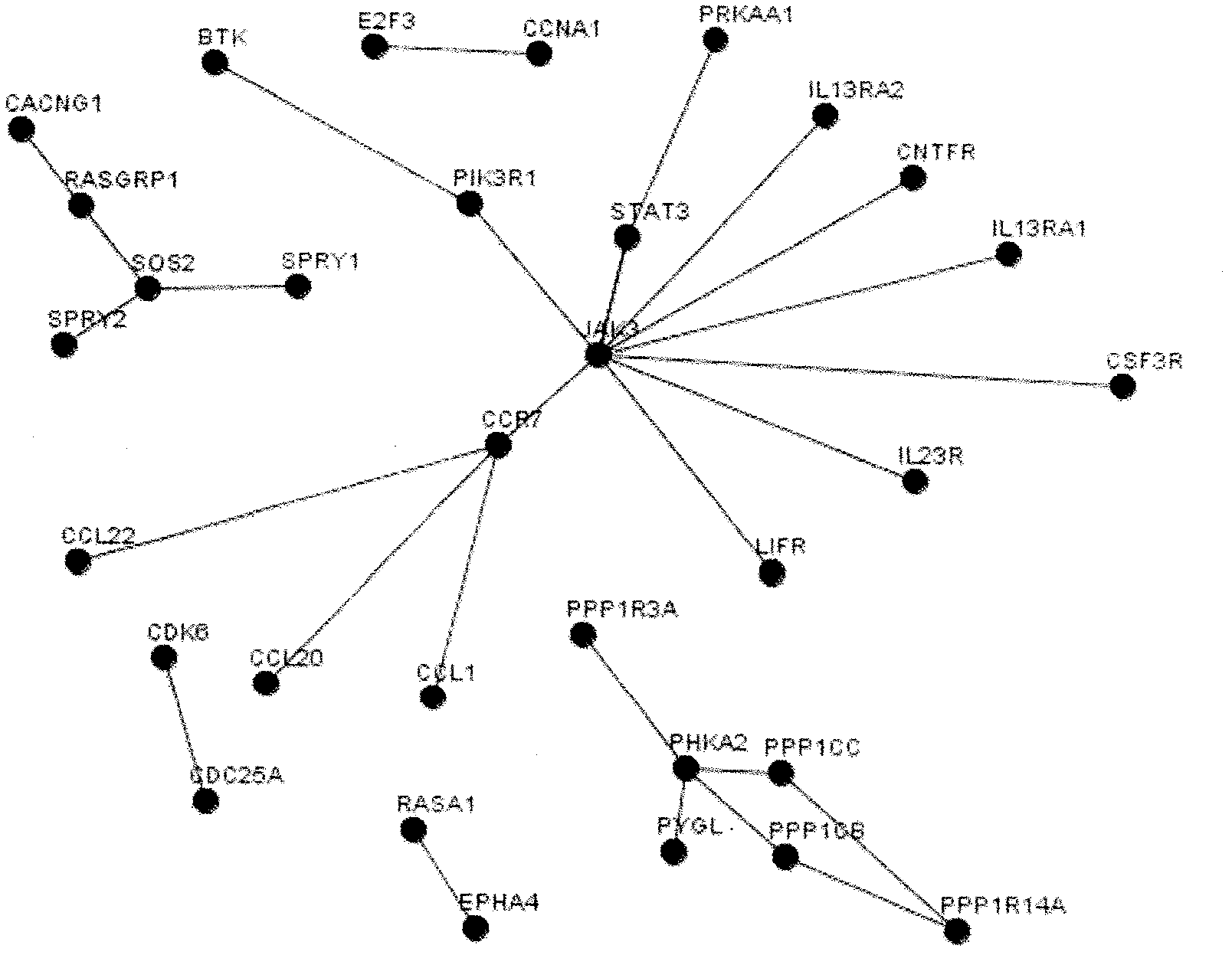
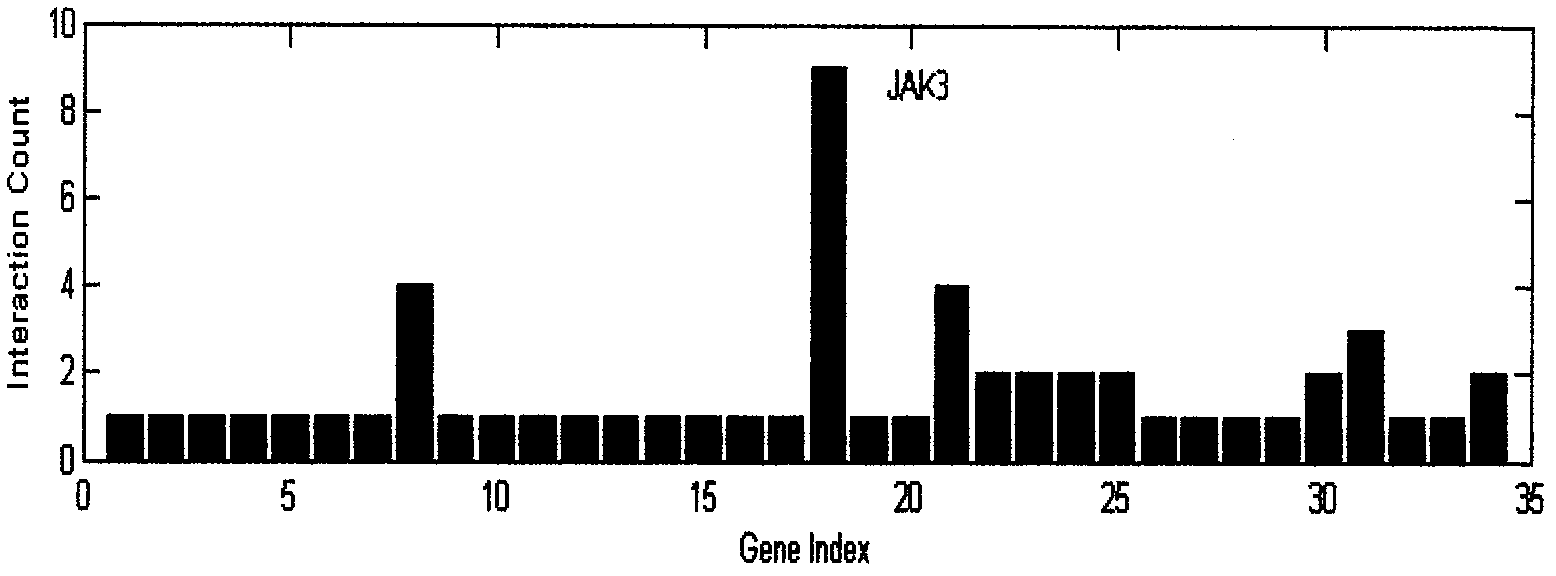
[0020] Step 2. Here we use our literature mining analysis system (software copyright registration number: 2009SR045504) to search and analyze miR-21 as the keyword, process the analysis results and extract the genes related to miR-21 we need, and organize them into list.
[0021] Step 3: Predict the target gene of miR-21. In this example, we will use the following three most commonly used software for forecasting:
[0022] PicTar 2005: http: / / pictar.mdc-berlin.de / cgi-bi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com