Human anti-human CD20 monoclonal antibody molecule and application thereof
A fully human and antibody technology, applied in the fields of application, anti-receptor/cell surface antigen/cell surface determinant immunoglobulin, antibody, etc., can solve the problem of unsatisfactory therapeutic agents
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Example 1. Construction of a large-scale fully human antibody library
[0075] This example describes how to construct a very large-scale fully human natural antibody library in Fab format. The Fab antibody library of the present invention was constructed by using blood samples from more than 3,000 healthy adults from different regions and different ethnicities, referring to the following documents, and the detailed construction process is described after the bibliography.
[0076] 1. Dantas, BC, et al, 2005, Construction of a human Fab phage display library from antibody repertoires of osteosarcoma patients. Gene. Mo. Res, 4(2): 126-140.
[0077] 2. Hiroshi, T, et al, 1999, Preparation of Recombinant Human Monoclonal Antibody Fab Fragments Specific for Entamoeba histolytica, Clinical and Diagnostic Labor Immunol, May 1999, 383-387.
[0078] 3. Wu, BP, et al, 2001, Construction and selection of the natural immune Fab antibody phage display library from patients with co...
Embodiment 2
[0097] Example 2 Using human CD20 to screen the fully human antibody library
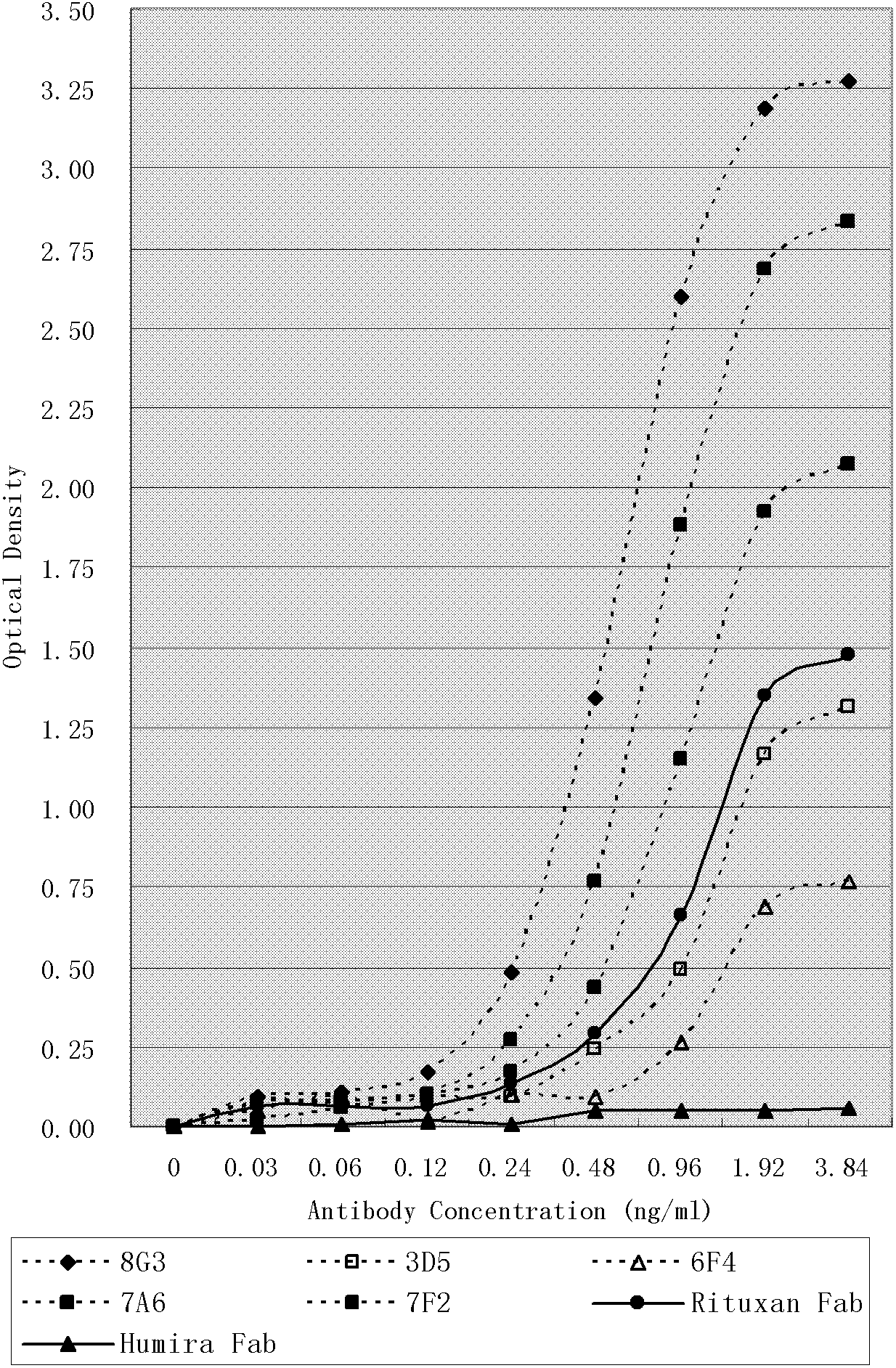
[0098] The self-prepared 2F2 (HuMax-CD20, GenMab) Fab was used as a positive control, and the recombinant human CD20 was used as an antigen to screen the above HuLib. The panning process is as follows:
[0099] Add 1 small portion of HuLib to a 25ml cell culture square bottle pre-coated with recombinant human CD20 protein, and incubate at 37°C for 1 hour. After washing 20 times with PBS containing 1% Tween-20, 1 ml of logarithmic phase TG1 cells were added, and cultured with shaking at 37° C. for 16 hours. Centrifuge at 12000rpm for 10 minutes, and transfer the supernatant to a new 50ml test tube. Take 500 μl of supernatant, and pan again according to the above method, for a total of 4 rounds. After the final round, the obtained bacterial suspension was diluted to 100,000 cells / ml and selected on 1.5% agar plates containing 0.1% ampicillin to obtain single spots. Inoculate ten 96-well deep-well ...
Embodiment 3
[0109] Example 3 Molecular Evolution
[0110] All CDRs of the 7F2 clone were analyzed by the method of Cunningham et al. (Cunningham and Wells, Science, 244:1081-1085, 1989), and residues sensitive to glycine substitution were identified, which were preferred mutation sites. The following table shows the preferred amino acid positions in the 7F2 clone, where the underlined ones are extremely sensitive to substitutions, and the dotted ones are relatively sensitive to substitutions, and they are all preferred mutation positions.
[0111]
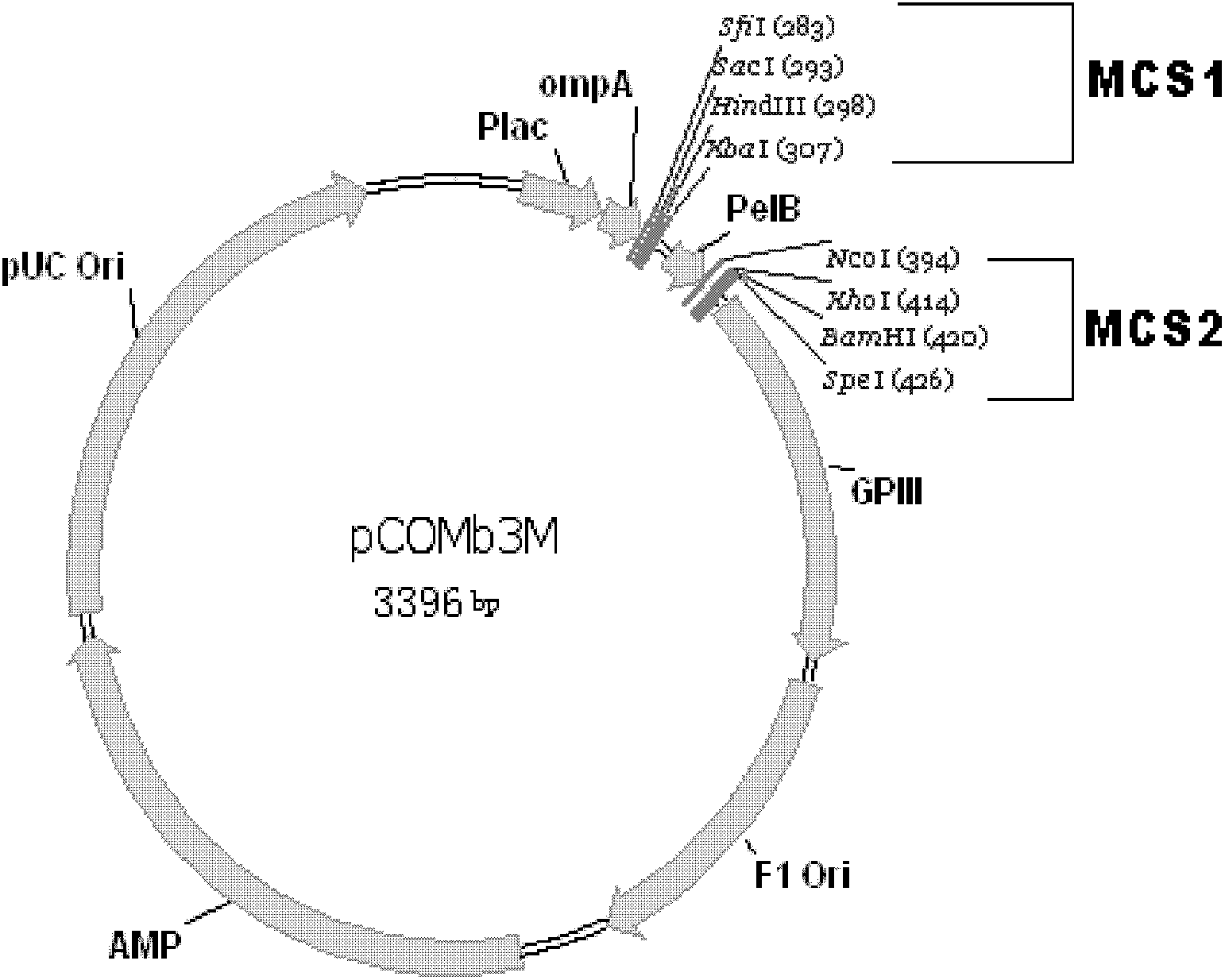
[0112] The above-mentioned preferred sites are mutated by oligonucleotide-mediated random mutation (Kunkel method), and further improved antibody molecules for these sites can be obtained by panning. The brief process is: in order to introduce mutations at the above preferred sites, mutation primers were designed according to the Kunkel method, and a Fab secondary antibody library was constructed using pCOMb3M as a vector. Using the method...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Dissociation constant | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com