Method for fast detecting low-frequency exogenous chromosome fragments of genes with SSR markers
A technology of markers and genetic markers, applied in the direction of biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of long-term, large manpower and material resources, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
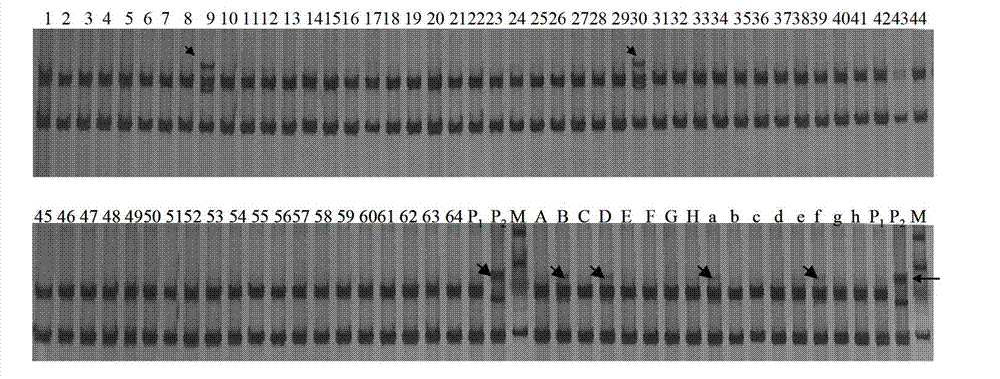
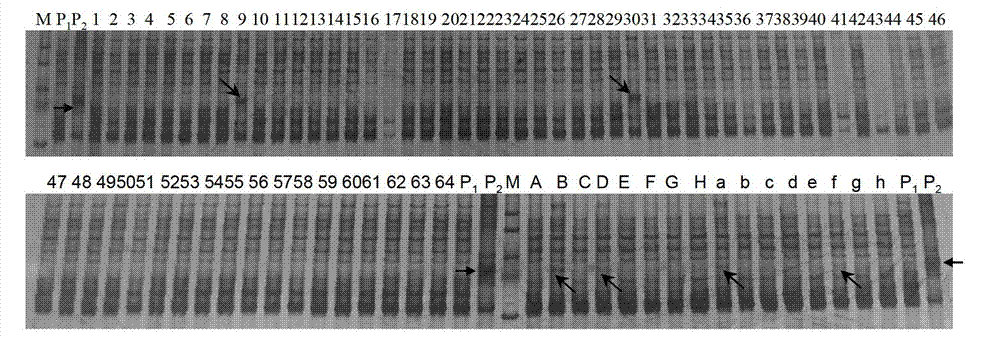
[0049] Example 1. Using SSR markers to quickly detect small fragments or genes of low-frequency exogenous chromosomes
[0050] In this embodiment, the method for rapidly detecting small fragments or genes of low-frequency exogenous chromosomes using SSR markers specifically includes the following steps:
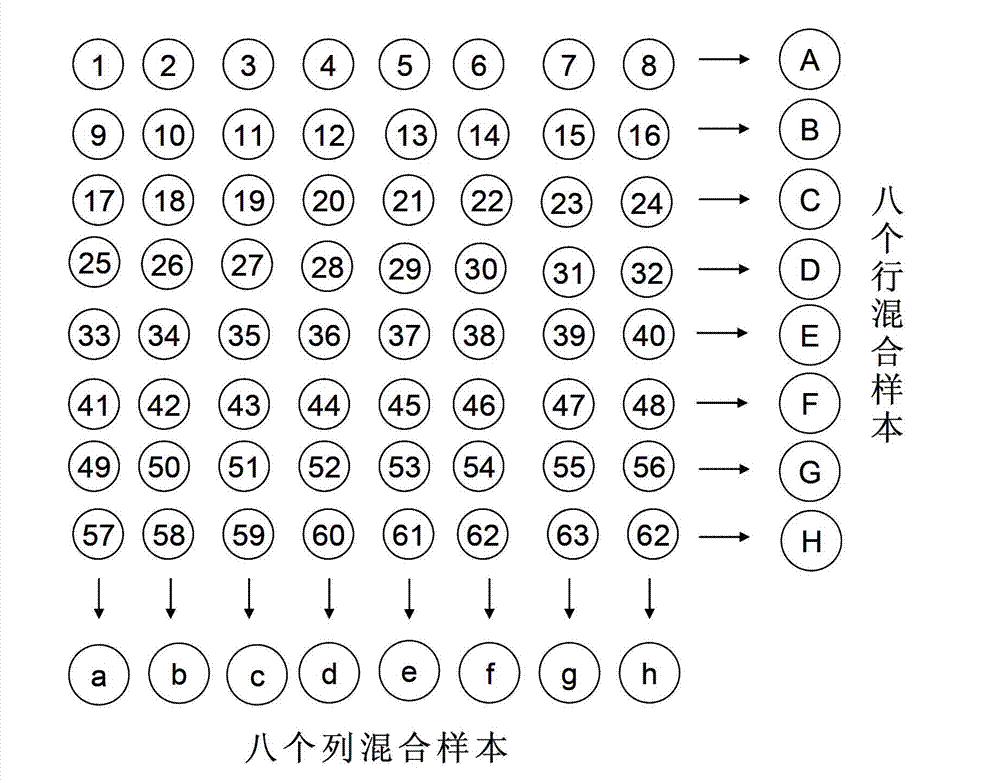
[0051] (a) Arrange the N samples to be tested according to the matrix A shown in formula (1);
[0052] A = a 11 a 12 · · · a 1 n a 21 a ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com