Neighbor-first Biomolecular Subnetwork Search Method Based on Expert Knowledge and Topological Similarity
An expert knowledge, biomolecular technology, applied in the computer field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0093] Preferred embodiments of the present invention will be further described in detail below in conjunction with the accompanying drawings.
[0094] In this embodiment, the experiment of the neighbor priority biomolecular subnet search method based on expert knowledge and topological similarity of the present invention is completed on a cluster computer of the Institute of Systems Biotechnology of Shanghai University. The cluster consists of 14 IBMHS21 blade servers and 2 The x3650 server forms computing and management nodes, and the network connection adopts Gigabit Ethernet and infiniband2.5G network. Each node is configured with two dual-core CPUs and 4GB of memory, each CPU is intelxeon51502.66GMhz main frequency, and two graphics workstations are used as front-end machines for scientific data visualization.
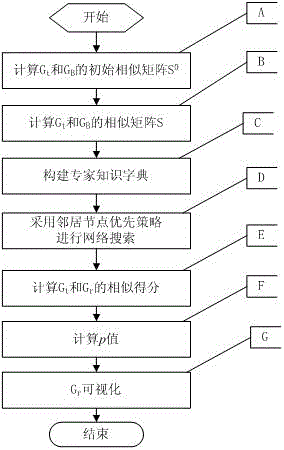
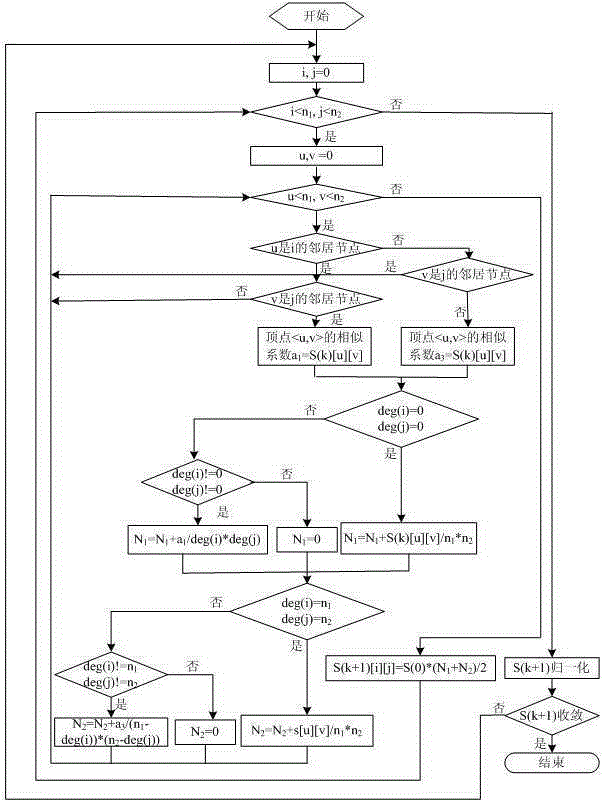
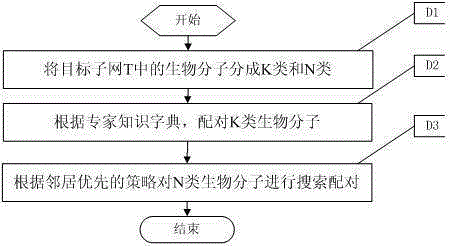
[0095] For network A (G A ), Network B (G B ) and Network A (G A ) in the target subnet T (G t ), the present invention's neighbor-first biomolecular subnet s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com