Protein-ligand affinity predicting method based on molecule descriptors
A molecular descriptor and affinity technology, applied in the field of protein-ligand affinity prediction based on molecular descriptors, can solve the problems of large target dependence, poor sensitivity of homologues, poor correlation, etc., and achieve strong predictive ability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
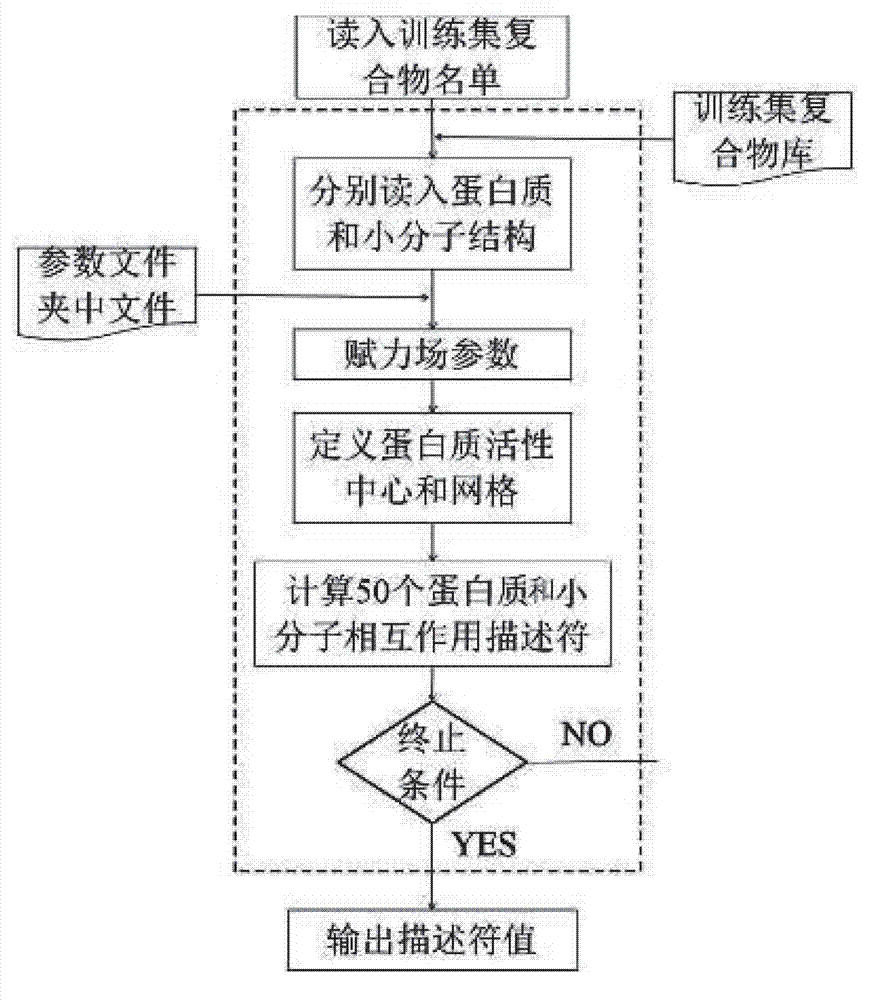
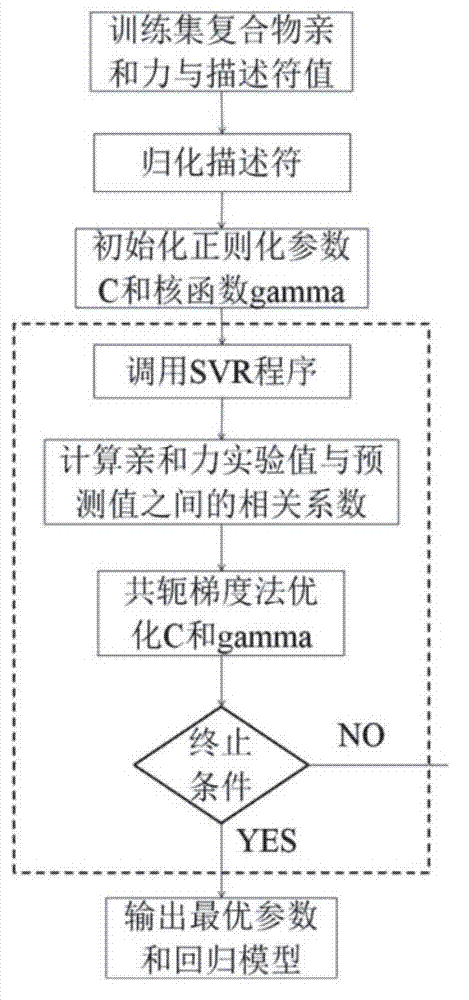
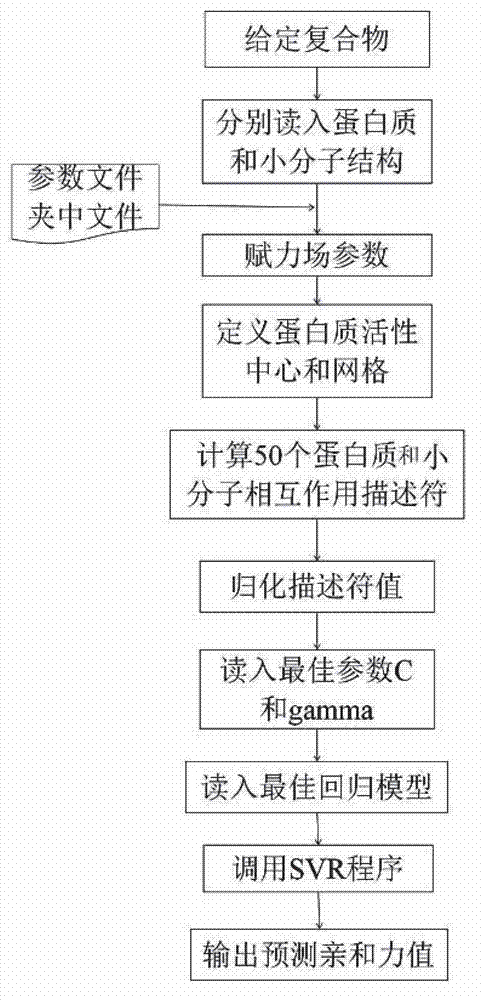
[0037] The method of the present invention belongs to the scoring method based on the empirical scoring function. By collecting 2278 diversified crystal structures of protein and ligand complexes and their binding affinity experimental values, 50 perfect and systematic molecules related to the interaction of proteins and ligands are constructed. Descriptors are used to reflect the affinity of the compound, and the relationship between the descriptor and the affinity of the compound is established by the method of support vector regression, so as to construct an empirical scoring function for predicting the affinity of a given compound.
[0038] Specific steps are as follows:
[0039] (1) Preparation of training set:
[0040] In total, the training set contains 2278 complex structures and their affinity data. The protein structure and the ligand structure in each complex are named with the PDB ID number and saved in the same folder. The protein structure is saved in the PDB fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com