New ovarian cancer biomarkers and targets
A technology for ovarian and uterine cancer, applied in the field of new ovarian cancer biomarkers and targets, which can solve the problems of inaccessibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0105] Example 1.0 - Identification of ARID1A mutations in ovarian cancer
[0106] Because CCC is genome stable 8,9 , which would be expected to have a restricted mutational profile and recurrent mutations, which would be evident by analysis of a small number of cases 24 .
[0107] The inventors used RNA-seq to decipher the transcriptomes of 17 ovarian clear cell carcinomas. Gene fusions and small intermediate deletions or insertions detected by methods described in recently published literature 25,33,34 , while SNV uses SNVmix, a recently published Bayesian mixture-based algorithm to detect 35 . The vast majority of SNVs are predicted to be rare germline variants rather than somatic mutations. Therefore, the inventors used the same approach that led to the identification of FOXL2 mutations in granulosa cell tumors 1 To identify genes that are recurrently mutated in CCC but not in unrelated cancer types. The inventors identified mutations in the ARID1A gene in six of se...
Embodiment 2
[0111] Example 2.0 - Identification of Other SWI / SNF Genes Mutated in Ovarian Cancer
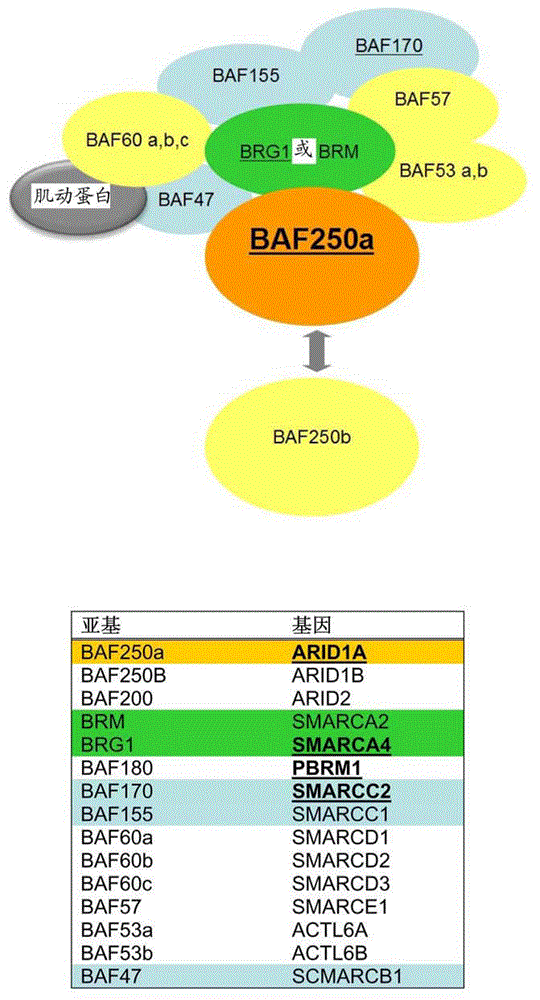
[0112] In ARID1A-mutation-negative CCC, the inventors identified SNVs in other SWI / SNFs, including missense mutations in SMARCA4 (encoding BRG1) and in PBRM1 (encoding BAF180) ( Figure 5 ). Since mutations in ARID1A or other SWI / SNF-encoding genes were seen in 9 of 18 CCCs (50%), these events were important for the development of this cancer. In contrast, mutations in TP53, BRAF, PIK3CA, and PTEN were only found in one, two, two, and three CCCs, respectively. Based on data from previously published literature, the proportion of cases predicted to carry these mutations 15 .
[0113] for figure 1 , the SWI / SNF complex components in which mutations were detected are highlighted. All 15 genes encoding protein components of the SWI / SNF complex are shown on the left of the table. Arrows indicate that either BAF250a or BAF250b may be present in the complex. The PBAF SWI / SNF complex (not sh...
Embodiment 3
[0115] Example 3.0 Mutations in ARID1A are associated with loss of BAF250a expression
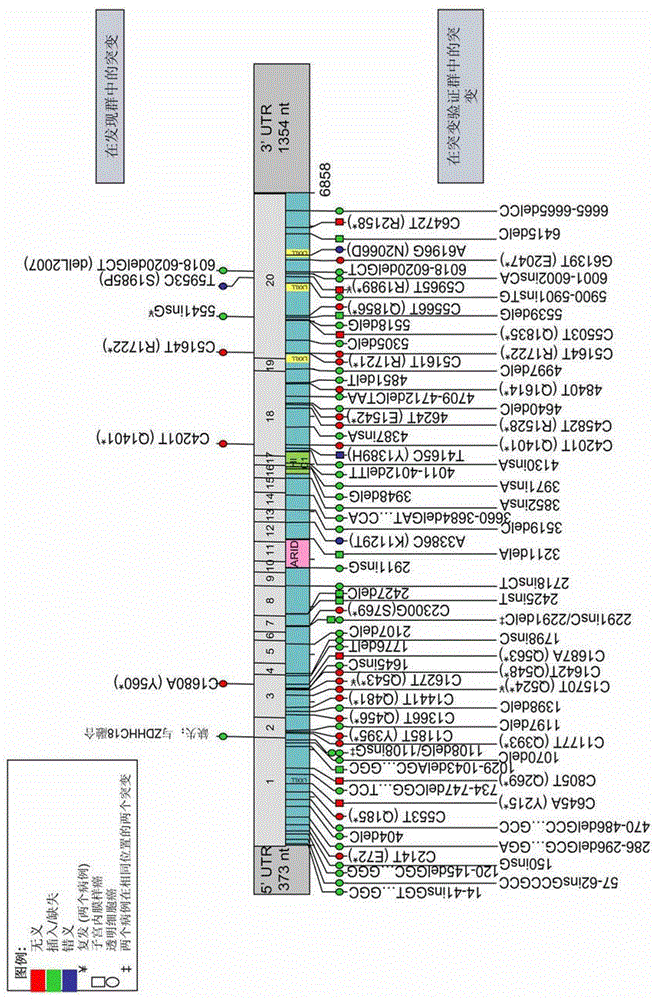
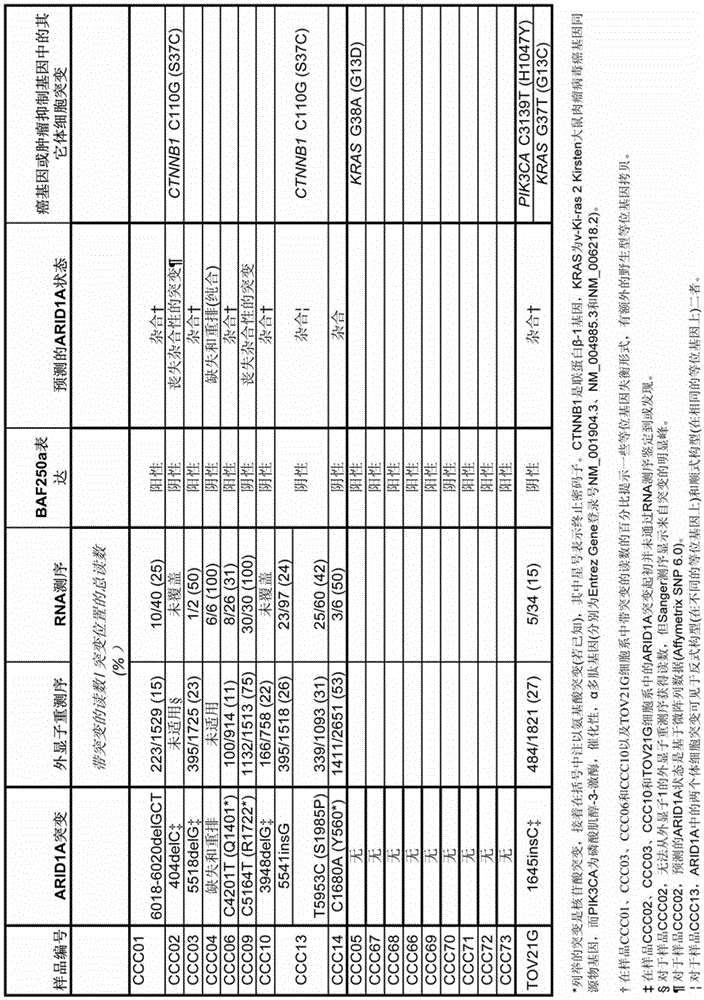
[0116] To elucidate that ARID1A mutations are associated with loss of expression, the inventors used a mouse monoclonal antibody (Abgent, Inc.) targeting the central region of the BAF250a protein. This antibody strongly stains all normal nuclei. Of the 18 clear cell carcinoma samples analyzed by RNA-seq in Example 1.0, eight showed loss of BAF250a expression. Of these eight cases, five had ARID1A mutations ( image 3 ). Interestingly, in the other three cases that stained negative for BAF250a by immunohistochemistry, no ARID1A mutation was detected by RNA-seq, suggesting that there may be other genetic or epigenetic mechanisms for the loss of BAF250a expression. Two cases with ARID1A mutations expressed BAF250a; one contained an in-frame deletion of a single amino acid in exon 20 (6018-6020delGCT(ΔL2007) in CCC01), while the other contained a premature termination in exon 18 SNV of the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com